Team:TU-Munich/Modeling/Protein Predictions
From 2013.igem.org
(→Prediction of Possible Structure using iTasser) |
(→Search for Homologous Structures using HHpred) |
||
| Line 101: | Line 101: | ||
|} | |} | ||
| - | + | ====Results:==== | |
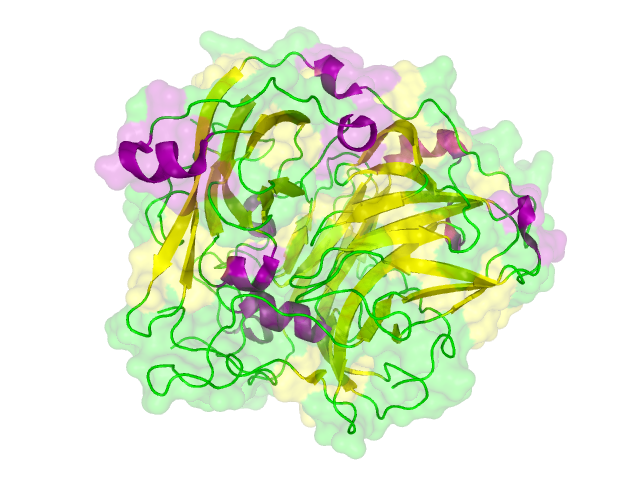
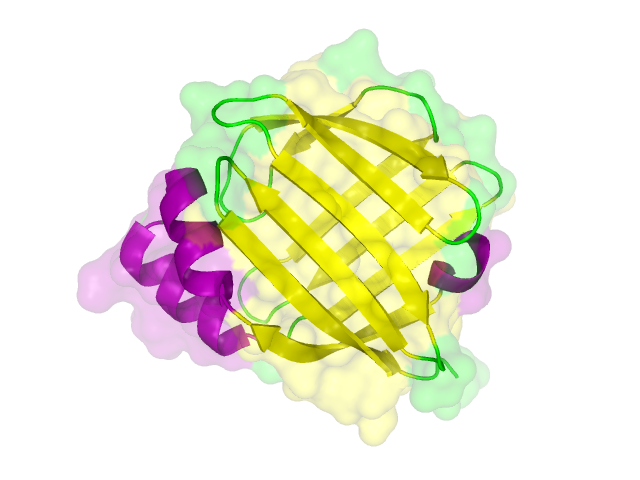
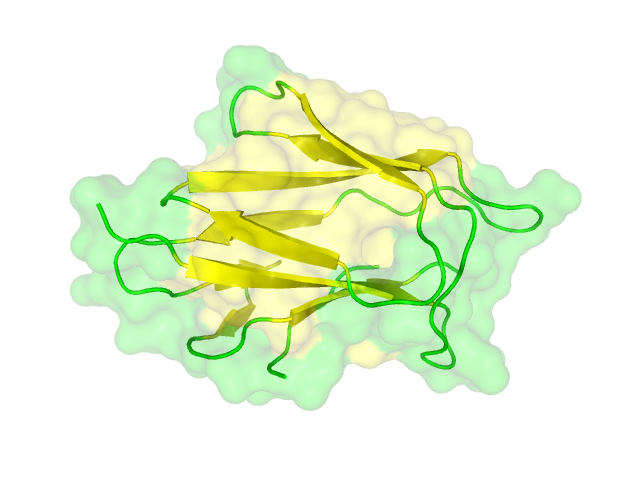
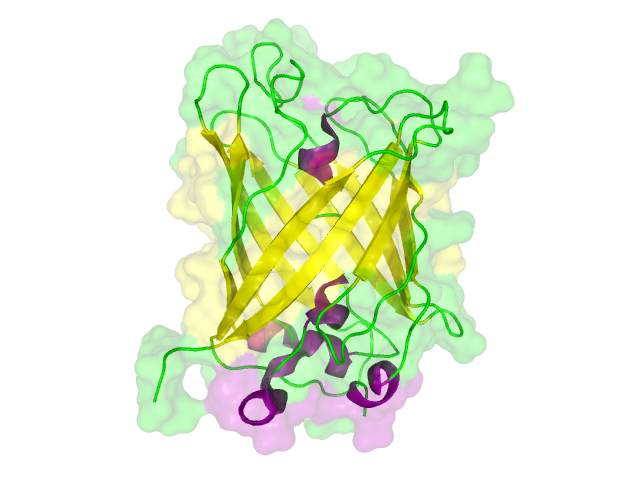
| - | + | The homology search showed that some effector proteins have some very close related proteins with a solved structure whereas for others no structure of a related protein has been solved so far. For example for the SypCatcher, PP1 and GFP very similar protein structures are availible which show and identitiy of above 90%. Some other effector proteins such as XylE, the laccase or the DDT Dehydrochlorinase have related protein wherefor the structure still gives good hints for structural questions on the effector proteins. For some other effector proteins there are only structures solved that show a very weak identity with our protein of interst wherefor just the rough fold can be expected to be in agreement for these proteins. Examples for such structurally unknown proteins are the NanoLuc which has been identified from shript this year and were not solved so far. Other expampls for structurally not determined proteins are the erythromycin esterase (EreB) and the transmembrane domain of the SERK receptor. <br> | |
| + | The structures obtained in this part were used for planing of of experiments, a homology modelling for the laccase was performed to investigate the probability of this BioBrick to contain disulphid bridges and most importantly the homologous structures were used as illustrations as it is shown in one of our [https://2013.igem.org/Team:TU-Munich/Results/How_To How-Tos]. | ||
==Analysis of Receptor Sequences – Choosing the right template == | ==Analysis of Receptor Sequences – Choosing the right template == | ||
Revision as of 21:38, 1 October 2013
Prediction of Protein Structures
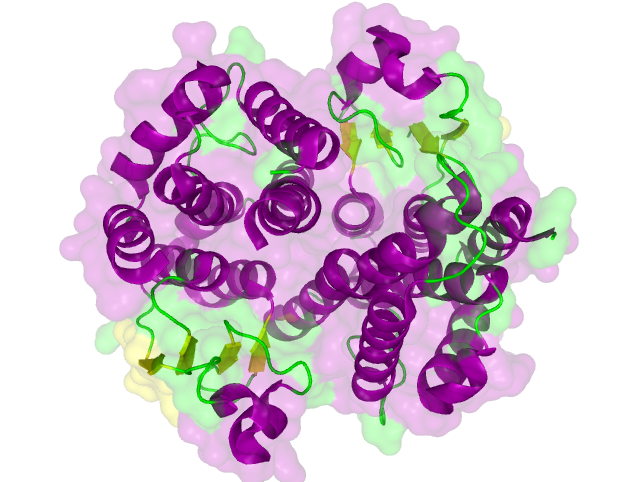
Structural propterties of effectorproteins are often important for their function and it is advantageous. For expamle it is necessary to know whether termini are accessible for protein fusion or whether the protein is functional in a multimeric fold. For this reason a search for structures was performed in the [http://www.rcsb.org/pdb/home/home.do protein data bank]. As the number of solved structures is still limited it is a promising attempt to search for homologous protein for which a cristall structure has been solved.
Search for Homologous Structures using HHpred
The search for homologous structures was performed using the free accessible webserver HHpred http://www.ncbi.nlm.nih.gov/pubmed/15980461 Söding et al., 2005. The protein sequence for the BioBricks was translated to amino acid sequeces using the AutoAnnotator and was then inserted into the the search field. The results are shown in table 1 for all proteins investigated in our project.
Results:
The homology search showed that some effector proteins have some very close related proteins with a solved structure whereas for others no structure of a related protein has been solved so far. For example for the SypCatcher, PP1 and GFP very similar protein structures are availible which show and identitiy of above 90%. Some other effector proteins such as XylE, the laccase or the DDT Dehydrochlorinase have related protein wherefor the structure still gives good hints for structural questions on the effector proteins. For some other effector proteins there are only structures solved that show a very weak identity with our protein of interst wherefor just the rough fold can be expected to be in agreement for these proteins. Examples for such structurally unknown proteins are the NanoLuc which has been identified from shript this year and were not solved so far. Other expampls for structurally not determined proteins are the erythromycin esterase (EreB) and the transmembrane domain of the SERK receptor.
The structures obtained in this part were used for planing of of experiments, a homology modelling for the laccase was performed to investigate the probability of this BioBrick to contain disulphid bridges and most importantly the homologous structures were used as illustrations as it is shown in one of our How-Tos.
Analysis of Receptor Sequences – Choosing the right template
For several purposes in our project we needed a synthetic receptor that enables us to express protein-domains at the cellular or extracellular side of the cell membrane. As a template we investigated several different plant-receptors form the well understood dicotyledon Arabidopsis thaliana and the moss we are currently using as our chassis Physcomitrella patens. The A. taliana-receptors have the advantage that their transgenic expression has successfully been demonstrated (Ref.) whereas the P. patens-receptors bear less risk that they do not function in the evolutionary distant moss (Ref).
As there were many different availible receptors that we could use as a template for our synthetic receptor we applied bioinformatical methods to evaluate the suitability of thes receptors. From this work the three examples ERF, FLS2 and SERK are shown (see Table 2).
| Receptor | Organism | Length (aa) | Sequence reference | Literature reference |
| ERF | A. thaliana | 1031 | [http://www.ncbi.nlm.nih.gov/protein/NP_197548.1 NP_197548.1] | |
| FLS2 | A. thaliana | 1173 | [http://www.ncbi.nlm.nih.gov/protein/NP_199445.1 NP_199445.1] | |
| SERK | P. patens | 625 | [http://www.ncbi.nlm.nih.gov/protein/XP_001759122.1 XP_001759122.1] | [http://www.freidok.uni-freiburg.de/volltexte/5390/pdf/Lienhart_Dissertation_2008.pdf Lienhart, 2007] |
Prediction of Signal Peptides
Introduction: The first analysis were performed to identify the signal-peptide that is bound by the cellular signal recognition particle and lead to the translocation of the polypeptide into the ER. The signal peptide becomes afterwards cleaved by a signal peptidase at a distict site. The analysis of the signal peptides was carried out using the [http://www.cbs.dtu.dk/services/SignalP SignalP 4.1 Server].
Results:
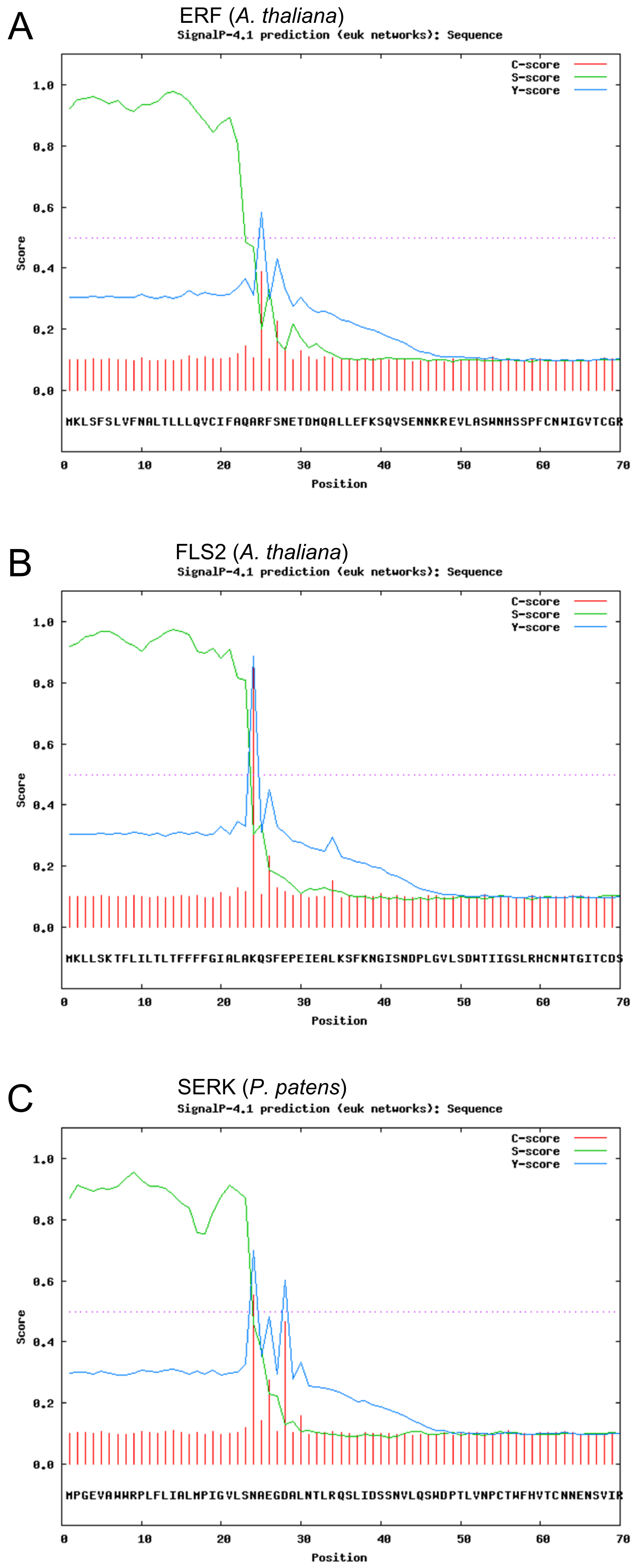
The prediction of the signal peptides was carried out for different receptors and will be illustrated for the three mentioned examples for which a signal peptide could be identified (see fig. 3).
The figure shows the N-terminal sequence of the receptors together with three scores: (1) the C-score (raw cleavage site score) in red, (2) the S-score (signal peptide score) in green and (3) the Y-score (combined cleavage site score) in blue.
The C-score shows the most probable cleavage site for the signal peptidase that could be identified for all shown receptors with a unclear result with two possible cleavage sites for the SERK-receptor. The amino acid with the highest C-score is according to the algorithm predicted to be the first amino acid of the cleaved receptor. The S-score was developed to identify amino acid sequences the lie in a polypeptide and others that do belong to the matured receptor. The course of this parameter is high for the first 23-28 amino acids for all receptors identifing these residues as signal peptides and decreases to low values quickly. The amino acid residue that lies at the greatest fall is the predicted border between the n-terminal signal peptide and the receptor. The Y-score as the third parameters represents the geometrical of the two previous parameters. It illustrates that the two first parameters show a good agreement for the identification of the signal peptide in all three illustrated receptors.
Discussion: It can be concluded that all three three depicted receptors seem to contain a sequence that functions as a signalpeptide. For many of the predicted receptors in the genome of P. patens this prediction did not yield a positive result. Concerning the signal peptide all mentioned receptors would be suitable as a template for our synthetic receptor. Although the SERK-receptor is favourable after this analysis as it's signal peptide is most probably recognized by the cellular machinery in P. patens and beers the smallest risk of failure.
Prediction of Transmembrane Regions
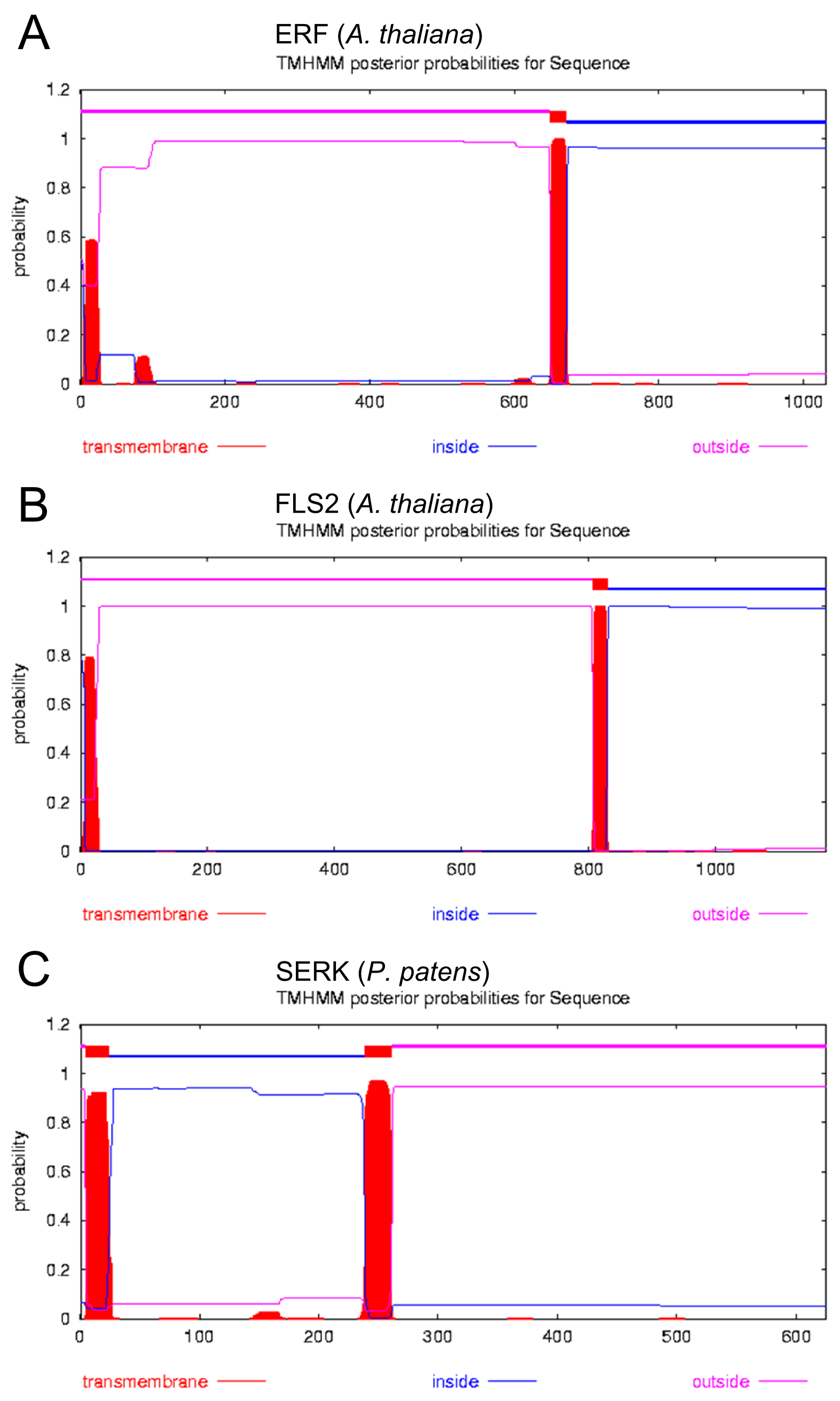
Introduction: Beside the identification of the signal peptide it was important to identify trans-membrane regions within the receptors as we wanted to use a type I receptor as a template that contains a N-terminal extracellular domain, a trans-domain region and a C-terminal intracellular domain. For this analysis the prediction tool [http://www.cbs.dtu.dk/services/TMHMM TMHMM] was applied for several different receptors and is again depicted for the reseptors ERK, FLS2 and SERK.
Results: The analysis yielded a singal peptide and a single trans membrane domain for all the depicted receptors (see fig. 4).
Discussion:
Prediction of Cellular Localization
Introduction: [http://wolfpsort.org Wolfpsort]
Results:
Discussion:
Choice of the SERK Receptor
<partinfo>BBa_K1159017</partinfo> <partinfo>BBa_K1159018</partinfo>
Analysis of Protein-Ligand Interaction
Text
Structure based prediction using PDBePISA
Text
References:
http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
- http://www.ncbi.nlm.nih.gov/pubmed/15980461 Söding et al., 2005 Söding J, Biegert A, Lupas AN. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W244-8.
- http://www.freidok.uni-freiburg.de/volltexte/5390/pdf/Lienhart_Dissertation_2008.pdf Lienhart, 2007 Lienhart Otmar. Untersuchungen zu einem Somatic-Embryogenesis-Receptor-like-Kinase-Homolog in Physcomitrella patens (Hedw.) B.S.G. PhD-thesis at Freiburg University
 "
"





AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351