Team:TU-Munich/Project/Bioaccumulation
From 2013.igem.org
BioAccumulation
BioAccumulation
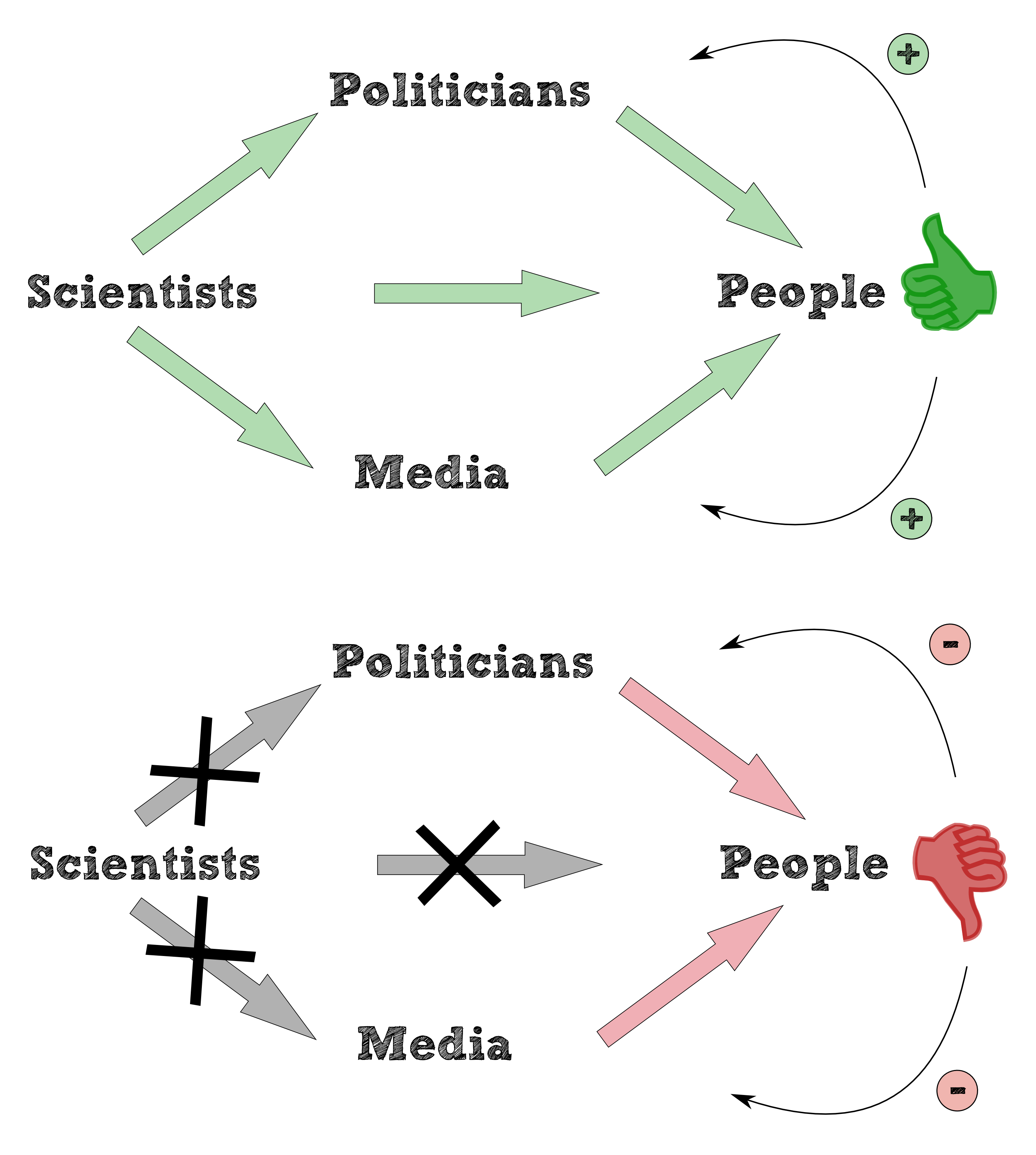
In general bioaccumulation is a versatile and ambiguous concept to be defined. “General term describing a process by which chemicals are taken up by an organism either directly from exposure to a contaminated medium or by consumption of food containing the chemical.” is the definition of bioaccumulation of the U.S. Environmental Protection Agency, 2010. The majority of definitions is aimed at the vicious circle of the predator-prey relationship caused by the bioaccumulation shows the build up of persistent chemicals such as DDT, PBC or dioxins in aquatic and terrestrial organisms. Health problems in humans, the survival of some affected populations and overall biodiversity in aquatic and terrestrial ecosystems are at risk and a result of the consume and the accumulation of persistent chemicals along the food chain (see figure 1 ). The Minamata catastrophe 1950 substantiates the result of mecury poisoning: the Chisso Corporation in Minamata released untreated effluent containing methylmercurychloride into Minamata Bay.
In Respect to Bioaccumulation we propose the employment of transgenic moos (Physcomitrella patens ) to produce effectors for enzymatic specific binding of pollutants additional to the biodegradation method for the development of a functional wastewater filter system which convert noxious critical substances in wastewater into non-hazardous compounds .Bioaccumulation is a possibility to remove xenobiotics from the environment by binding them to a protein that has been designed for this purpose (see figure 2). There are a broad range of natural as well as engineered binding proteins available. Natural binding proteins act as a model and initiator in design of new artificial binding proteins regarding research in different fields of biotechnology. Lipocalins, natural binding proteins, as base and scaffold for the design of anticalins confirms the popularity of bioaccumulated proteins in red biotechnology . The role of Fibrillin in the abscisic acid-mediated photoprotection shows an example of functioned bioaccumulation in a plant . The most commonly known binding proteins are antibodies which defend mammals against pathogens and toxins. Beside these natural binding proteins and anticalins there are more and more designed binding proteins such as Affibodies derived from the z-domain of the antibody-binding protein A (Ref) and DARPins that are based on an ankyrin scaffold .Critical substances such as Bisphenol A known as an endocrine disrupter which can mimic endrogen and the anti-inflammatory drug used as analgesic Diclofenac could accumulate by our physco filter. The last effects in contrast to the negative health effects developed from Bisphenol A ecological effects like on freshwater fish species. The localization of the effectors is exclusive membrane associated in contrast to the cytoplasmatic or secretory localization by biodegraded effectors.
The Fluorescein binding Anticalin FluA
Mostly in biochemical research applications a synthetic organic dark red powder is used to label and track cells as well as target to specific proteins or structures within cells. Stunningly that this red coloured powder was the reason for the green dye of the Chicago River on St. Patrick’s Day on Ireland in 1962 . Meant is the well-known fluorophore fluorescein which was used from the iGEM team Freiburg 2008 to demonstrate the high expression level of the fusion protein and the localization at the cell membrane obvious by the strong fluorescent signal at the cell surface for their Modular Synthetic Receptor System. To develop its fluorescence character an artificial designed binding protein was necessary. The choice fell on the lipocalin fluA which variants of the fluorescein binding confirms table 1. To prove the extracellular membranbinding localization of our constructs in context of bioremediation the interaction between fluA and flourescein was chosen . In contrast to iGEM Freiburg we used another biobrick for flu A based on the necessarily of higher dissociation constant for our Physco filter system (see table 1). So we created a transgenic moss plants (PF-15) with the fluorescein binding anticalin flu A on the extracellular part of the receptor . Producing engineered proteins for medical purpose in the red biotechnology is one of the main research fields in which anticalins play a major role. Lipocalins such as bilin-binding-protein BBP (from Pieris brassicae) can be used for generating molecular pockets with a diversity of shapes and for creating a stable receptor protein for a ligand of choice, so that development of binding proteins against nearly chemical structures with comparable size is possible. Figure 3 shows the principle of reshape the binding pocket by amino acid substitutions in context of the conversion from BBP to flu A. The binding site of the αβ-scaffold of BBP is formed by four loops on the top of an eight-standed β-barrel. To recognize fluorescein instead of bilin 16 residues in the binding center were encountered a random mutagenesis. Ligand binding studies and mutagenesis experiments shown the specificity of the molecular recognition of fluorescein through hydrophobic packing, charged sidechain environment and hydrogen bonds with its hydroxyl- groups, the responsibility for tight complex formation by charged residues at the pocket center and the variability of the randomizes amino acid positions. The specificity is encoded in the first shell residues of the binding pocket shows the following publication in which the swap of binding specificity happened via binding pocket grafting . In addition to the changed loop region which leads to a deeper cavity of fluorescein than bilin the conformation of the base of the binding pocket shows a rearrangement. The electron transfer can be explained with the interaction between Trp 129 with the xanthenolone part .
Table 1:
Variants of the fluorescein binding Anticalin FluA | |||
| Proteinvariant | KD of FluA to fluorescein | Literature reference | BioBrick |
| FluA | 152 nM | http://www.ncbi.nlm.nih.gov/pubmed/10051566 Beste et al., 1999 | <partinfo>BBa_K157004</partinfo> |
| FluA (R95K) | 64 nM | http://www.ncbi.nlm.nih.gov/pubmed/16307475 Vopel et al., 2005 | not availible as BioBrick |
| FluA (R95K, A45I, S114T) | 2 nM | http://www.ncbi.nlm.nih.gov/pubmed/16307475 Vopel et al., 2005 | <partinfo>BBa_K1159002</partinfo> |
Glutathione S-transferase
- General principle of glutathion conjugation
- Trap mechanism: chemical compound is much more soluble after conjugation and becomes transported into the vacuole.
- Already a natural mechanism in plants. Enzymes might be limited and genetic engineering can focuss this existing problem on specific chemical compounds.
- Transgenic moss created
- Usage of an existing BioBrick <partinfo>BBa_K620000</partinfo>
- Assay is the conjugation of Glutathion to Monochlorobimane which becomes fluorescent after conjugation.
Protein Phosphotase 1 - A molecular mop for Microcystin
[[File:TUM13 Physco-lifecycle.png|thumb|right|350px| Figure 1: Algenblüte, Microcystin, PP1 und Microcystein]
- From a collaboration with Dundee iGEM Team
- Enzyme (phosphatase) derived
Text
References:
- http://www.ncbi.nlm.nih.gov/pubmed/10051566 Beste et al., 1999 Beste G, Schmidt FS, Stibora T, Skerra A. (1999) Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. PNAS, 96(5):1898-903.
- http://www.ncbi.nlm.nih.gov/pubmed/16307475 Vopel et al., 2005 Vopel S, Mühlbach H, Skerra A. (2005) Rational engineering of a fluorescein-binding anticalin for improved ligand affinity. Biol. Chem., 386(11):1097-104.
- http://www.ncbi.nlm.nih.gov/pubmed/9255793 Nord et al., 1997 Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren PA. (1997) Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nature Biotech. 15(8):772-7.
 "
"



AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351