As there are no genes that can react to parathion specially, we need to degrade parathion firstly and then using biosensor to sense its product p-nitrophenol (PNP). The bacterial organophosphorus hydrolase enzyme hydrolyzes and detoxifies a broad range of toxic organophosphate pesticides by cleaving the various phosphorus-ester bonds. In the first part, we needed to use these enzymes to degrade parathion into PNP.

As we search the registry of parts, we found a part that is related to our project: BBa_K215090 (OpdA). But we hoped to obtain various enzymes and display them onto the membrane using ice nucleation protein to construct an effective degradation system, so we decided to test another organophosphorus hydrolysis from Flavobacterium sp.
PNP has a strong absorbance at 397nm. Naturally, absorbance at 397nm can be measured to test the efficiencies of parathion degradation.At first, a standard curve which reflected the relationship between PNP concentration and absorbance at 397nm was required and we quickly drew it by setting a PNP concentration gradient and testing their absorbance.
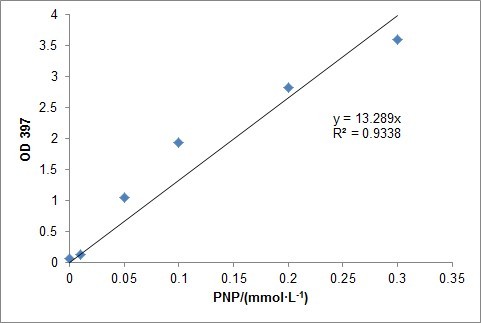
And then, we transformed opd into E.coli and tested their function. As our expectation, the result showed that under the function of these gene, parathion successfully degraded into PNP and DEP.

However,to standardize opd gene, we must mutate three sites. We do not have enough experiment time, so we have not standardize this part yet.
 "
"

