Team:TU-Eindhoven/IntegralModel
From 2013.igem.org
Pascalaldo (Talk | contribs) (→Merging the Models) |
Pascalaldo (Talk | contribs) (→Merging the Models) |
||
| Line 13: | Line 13: | ||
{{:Team:TU-Eindhoven/Template:Image | filename=IntegralModel.jpg}} | {{:Team:TU-Eindhoven/Template:Image | filename=IntegralModel.jpg}} | ||
{{:Team:TU-Eindhoven/Template:FloatEnd | caption=Schematic of the integral model | id=IntegralModelSchematics }} | {{:Team:TU-Eindhoven/Template:FloatEnd | caption=Schematic of the integral model | id=IntegralModelSchematics }} | ||
| + | |||
| + | This model was implemented as both a ODE and a stochastic model. | ||
==References== | ==References== | ||
Revision as of 15:20, 21 September 2013



Integral Model
The last step in creating a comprehensive model of the contrast mechanism is merging all individual models. Previously the model to predict the FNR concentration in a cell was described. This model will be combined with the decoy sites model, which was extended to contain the specific promotor that was used in this research. Here, the properties of the merged model will be discussed.
Merging the Models
In order to merge the FNR model and the decoy sites model, the exact connection between the two models had to be defined. The FNR model describes the (active) FNR concentration in the bacterial cells. This active FNR species fulfills the role of transcription factor in the decoy sites model. Using this information, the models could simply be combined by merging the Active FNR species of the FNR model and the T (transcription factor) of the decoy sites model. The resulting model schematics are depicted in .
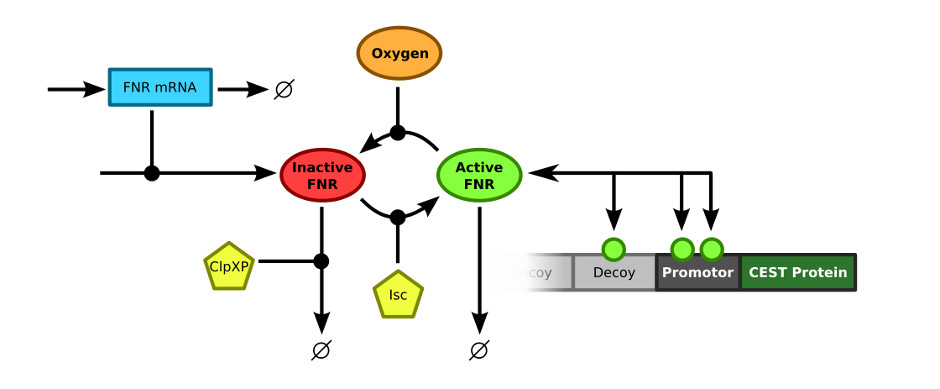
This model was implemented as both a ODE and a stochastic model.
References
 "
"



