Team:NYMU-Taipei/Project/Kill
From 2013.igem.org
| Line 20: | Line 20: | ||
[[image:NYMU_NO7_ethanol-producing.png]] | [[image:NYMU_NO7_ethanol-producing.png]] | ||
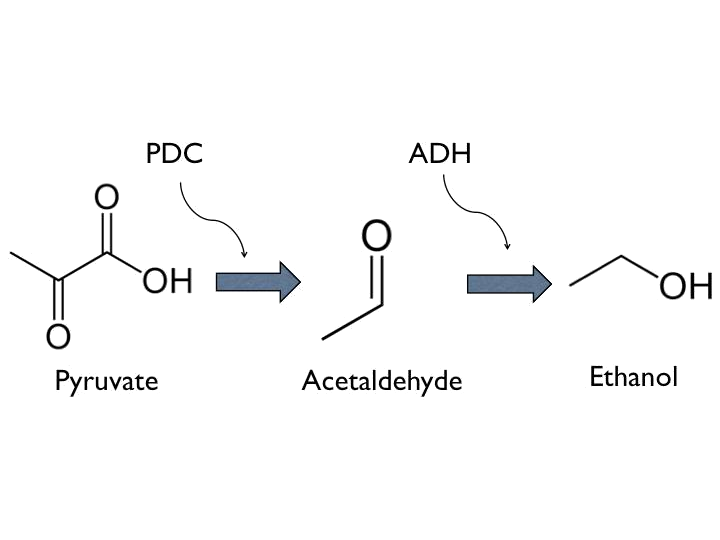
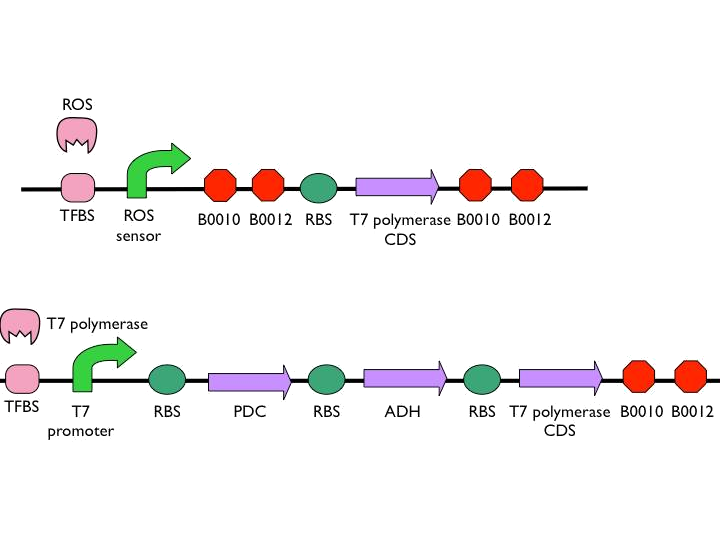
*The coding sequence of the first train is [http://parts.igem.org/Part:BBa_K145001 Part:BBa_K145001], which is the upstream DNA sequence of T7 polymerase. Otherwise, one of the coding sequence on the second strain will also be transcribed into the mRNA of T7 polymerase. In order to construct positive feedback of the alcohol-producing enzymes, we set a T7 polymerase coding sequence on the second train. Meanwhile in front of the second train, we place the receptor of T7 polymerase and [http://parts.igem.org/Part:BBa_I712074 T7 promoter]. In case of the leaky condition, we choose T7 polymerase as our go-between of alcohol-producing enzymes proliferating system. | *The coding sequence of the first train is [http://parts.igem.org/Part:BBa_K145001 Part:BBa_K145001], which is the upstream DNA sequence of T7 polymerase. Otherwise, one of the coding sequence on the second strain will also be transcribed into the mRNA of T7 polymerase. In order to construct positive feedback of the alcohol-producing enzymes, we set a T7 polymerase coding sequence on the second train. Meanwhile in front of the second train, we place the receptor of T7 polymerase and [http://parts.igem.org/Part:BBa_I712074 T7 promoter]. In case of the leaky condition, we choose T7 polymerase as our go-between of alcohol-producing enzymes proliferating system. | ||
| - | [[ | + | [[File:NYMU_no7_circuit.png|300px|''' Suicide Circuit''']] |
| - | + | ||
==<font color=darkorange>'''Experiment'''</font>== | ==<font color=darkorange>'''Experiment'''</font>== | ||
===Test the producing efficiency with positive feedback=== | ===Test the producing efficiency with positive feedback=== | ||
Revision as of 19:15, 27 September 2013


Contents |
Project overview
Introduction
Our aim is to resolve collapse colony disorder, which was characterized in recent years. However, it is a quite up-to-date disease, researchers have found some suspicious candidates resulting CCD. Our project focuses on one of main pathogens, Nosema ceranae. As the previous group stated, we used theoretically plausible sensor to sense Nosema ceranae, molecules to prevent Nosema ceranae from invading, and even molecules to kill Nosema ceranae. In case of uselessness of previous circuit on Nosema ceranae, we have to come up with another way to hold back the spreading of Nosema ceranae. This is what we concentrate on.
Background
- Ethanol, is known as a kind of toxins of bees. Up to certain degree, it have potential harm to the nervous system of bees. Besides, it could have influence on hydrostatical pressure, which is one of essential factors to the germination of Nosema ceranae.
- Ethanol can be produced from pyruvate through [http://parts.igem.org/Part:BBa_K1104400 Pyruvate Decarboxylase (PDC)] and [http://parts.igem.org/Part:BBa_K1104401 Alcohol Dehydrogenase (ADH)].
Method and circuit design
Method
The infected honeybee can go back to their hive, contact other bees, and spread the pathogen in a very short time. To prevent the whole bees in hive from infected,we come up with a way to prohibit Nosema from speading. The method is composed of two parts.
- 1.How to post-sensing if Bee. coli could not successfully kill the Nosema.
We use the ROS promoter from the previous circuit. What difference between our circuit and previous circuit is we directly put two terminators after the ROS promoter to construct threshold. In case our circuit will be easily activated to cause unnecessary toll.
- 2.How to suppress the spreading of Nosema if Bee. coli failed
Inhibit the extrusion of the polar filament or let it germinate at the wrong place, and thus the spores cannot get into the intestine cell. Since the mechanism that triggers the polar filament’s extrusion is hydrostatic pressure, ethanol is our potential candidate for it is able to reduce water permeation across cell membrane and phospholipid layers, also caused a powerful inhibition of spore germination. (In this way, Nosema senses much lower hydrostatic presure).
Circuit design
- According to the result of modeling, it is obvious that we should put 1~2 terminators whose forward efficiency is about 0.98 CC after our hybrid promoter. The purpose of the terminators is to prevent bees from dying easily.The purpose of the terminators is to prevent bees from dying easily. On top of that, the other important part is the positive feedback circuit. It is useless that we only produce very few pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH) to synthesize much less ethanol. Hence we introduced some biobricks to our circuit to exert efficiency.
- The coding sequence of the first train is [http://parts.igem.org/Part:BBa_K145001 Part:BBa_K145001], which is the upstream DNA sequence of T7 polymerase. Otherwise, one of the coding sequence on the second strain will also be transcribed into the mRNA of T7 polymerase. In order to construct positive feedback of the alcohol-producing enzymes, we set a T7 polymerase coding sequence on the second train. Meanwhile in front of the second train, we place the receptor of T7 polymerase and [http://parts.igem.org/Part:BBa_I712074 T7 promoter]. In case of the leaky condition, we choose T7 polymerase as our go-between of alcohol-producing enzymes proliferating system.
Experiment
Test the producing efficiency with positive feedback
Introduction
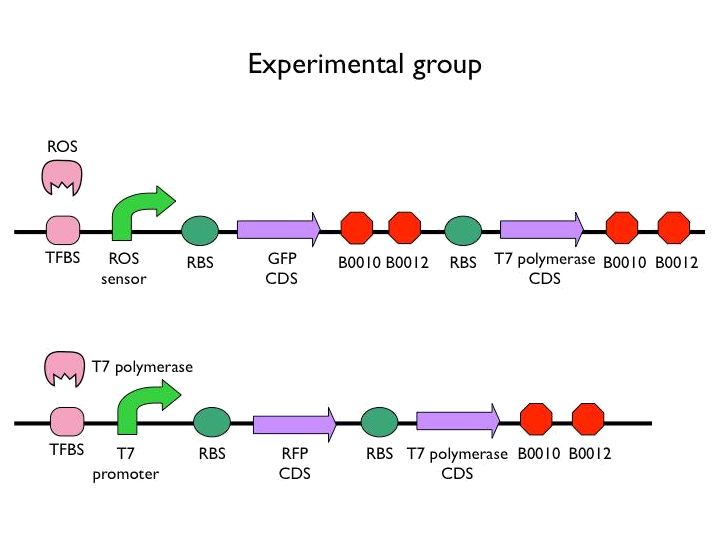
- It is not enough to just produce enzymes to synthesize alcohol. We need a more abrupt, rapid way to produce PDC and ADH which were enzymes responsible for synthesizing alcohol, so we add T7 system on the original circuit to construct positive feedback.
Background
- According to the experiment made by 5th group, we could know the efficiency of ROS promoter
- According to the experiment made by Caitlin Conboy, we could know the forward efficiency of B0015[http://parts.igem.org/Help:Terminators/Measurement/Caitlin_Conboy]
Material
- [http://parts.igem.org/Part:BBa_K1104205 ROS sensing system]
- T7 system
- [http://parts.igem.org/Part:BBa_B0015 B0015]
- [http://parts.igem.org/Part:BBa_E0840 GFP generator]
- [http://parts.igem.org/Part:BBa_I13507 RFP generator]
- H2O2
Outline
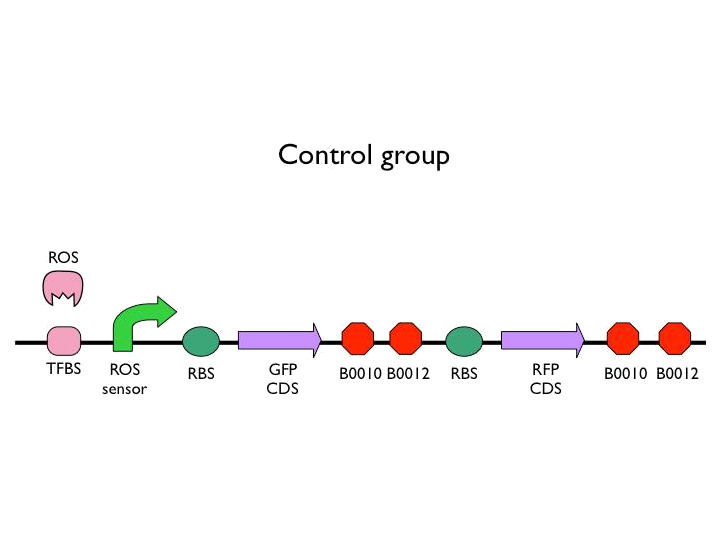
We created the circuit below. Every experiments are added the same concentration of H2O2. With reporter succeeding to the promoter, we could figure out whether the result was caused by promoter or not. And then there is the terminator we would test. After the terminator is the T7 polymerase which would open up the subsequent positive feedback. Besides, we used the other fluorescence protein as our second reporter. By testing the fluorescence from the second reporter, we could know how much proteins would be expressed on this site. Our control is the similar circuit without positive feedback below.
Method
Before the experiment, we placed 1 ul bacteria liquid into 2 ml LB and put them into incubator for experiment on the next day. After growing the bacteria, we sucked a few liquid we cultured before, testing the OD value respectively. If the OD value is lower than 0.5, put the liquid into incubator again, repeating the step until the OD value is up to 0.5. In every well, we add mixture of M9 buffer and H2O2. The concentration of H2O2 is 1mM where is the best range where ROS promoter could switch on. Then we put control and experiment sample which have been cultured overnight and have the same OD value into wells. Then measure the OD and fluorescence value of samples through thermo varioskan flash. After using thermo varioskan flash , put 96 wells back to incubator for another 1 hour. Repeat the measuring and culturing steps for 10 points. At last, analyze the data with wavelength of green and red light.
Test the lethal dose of ethanol to honey bees
Introduction
In the midgut of the bees, where filled with ethanol, there is a big problem that whether Bee. coli could survive the ethanol before the bee died. To find the answer, we have to understand the persistence of the bee to ethanol.
Background
- Harnessed honey bees readily consume 1%, 5%, 10%, and 20% ethanol solutons.
- 95% ethanol will also be consumed as long as the antennae do not make contact with the solution.
- With the exception of 95% ethanol, consumption as measured by contact time or amount consumed does not differ in animals that consume 1%, 5%, 10%, and 20% ethanol solutions.
- Exposure to a lesser (or greater) concentration of ethanol does not influence consumption of a greater (or lesser) concentration.[http://www.alcoholism-cer.com/pt/re/alcoholism/abstract.00000374-200008000-00004.htm;jsessionid=SFFFMpK8gJJJJgpnHN4T2bGnS1NjQh1rx0pNc7mh0n5MfpJp7hBV!1424873047!181195629!8091!-1]
- Total body fluid inside one bee: 33 uL (ave.)[http://www.jbc.org/content/66/1/77.full.pdf]
- Minimum inhibitory concentration for E.coli 2.73 uL / in 33uL body fluid[http://www.scielo.br/pdf/bjps/v45n2/v45n2a08.pdf]
Material
- Honey bees
- Sucrose solution
- EtOH 95%, density=0.8
- Color paintings, red and green
Method
- Paper survey, finding out EtOH lethal dose and sub-lethal dose for Bees
- Prepare EtOH sucrose solution:
- Lethal dose, 40 uL 95% EtOH dissolved in 360 uL sucrose solution and feed 20 uL/bee
- Sub-lethal dose, 20L 95% EtOH dissolved in 380 uL sucrose solution and feed 20 uL/bee
- Normal control, 400 uL sucrose solution and feed 20 uL/bee
- Cooling bees by putting them into 4°C refrigerator
- Feeding bees with different conc. EtOH sucrose solution by 200uL tips
- Marker bees with different color painting, lethal dose: red, sub-lethal dose: green, normal control: non
- Put bees into bee hives and begin color run for 10 min
- Observation and record
| Bee's color run | Result of bee's color run |
|---|---|

| 
|
Result
2 uL ethanol is enough to kill the bees. Comparing to Bee. coli, who need at least 2.73 uL ethanol to inhibit growth, honey bees are more fragile than Bee. coli.
Reference
- Mazzola, P. G., Jozala, A. F., Novaes, L. C. L., Moriel, P., & Penna, T. C. V. (June 01, 2009). Minimal inhibitory concentration (MIC) determination of disinfectant and/or sterilizing agents. Brazilian Journal of Pharmaceutical Sciences, 45, 2, 241-248.[http://www.jbc.org/content/66/1/77.full.pdf]
- Bishop, George H. (2011). Body fluid of the honey bee larva - II. Chemical constituents of the blood, and their osmotics effects.[http://www.scielo.br/pdf/bjps/v45n2/v45n2a08.pdf]
- Abramson, C. I., Stone, S. M., Ortez, R. A., Luccardi, A., Vann, K. L., Hanig, K. D., & Rice, J. (January 01, 2000). Neurobiological, Psychosocial, and Developmental Correlates of Drinking - The Development of an Ethanol Model Using Social Insects I: Behavior Studies of the Honey Bee (Apis mellifera L.). Alcoholism: Clinical and Experimental Research, 24, 8, 1153.[http://www.alcoholism-cer.com/pt/re/alcoholism/abstract.00000374-200008000-00004.htm;jsessionid=SDQRC8TrDPbvHJMqlm5qd7QQG3hvpyn7whPnqQ7LG2YPnQpDCQfs!1424873047!181195629!8091!-1]
- Higes, M., García-Palencia, P., Martín-Hernández, R., & Meana, A. (March 01, 2007). Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). Journal of Invertebrate Pathology, 94, 3, 211-217.[http://www.ncbi.nlm.nih.gov/pubmed/17217954]
- FRIXIONE, E. U. G. E. N. I. O., RUIZ, L. O. U. R. D. E. S., CERBÓN, J. O. R. G. E., & UNDEEN, A. L. B. E. R. T. H. (March 01, 1997). Germination of Nosema algerae (Microspora) Spores: Conditional Inhibition by D2O, Ethanol and Hg2+ Suggests Dependence of Water Influx upon Membrane Hydration and Specific Transmembrane Pathways. The Journal of Eukaryotic Microbiology, 44, 2, 109-116.[http://onlinelibrary.wiley.com/doi/10.1111/j.1550-7408.1997.tb05946.x/full]
- Registry of standard biological parts, BBa_K558001.[http://igem.ym.edu.tw/pv2013/index.php/Team:NYMU-Taipei/Nosema_spread_prevention#Method]
- 2010igem.UT-Tokyo, Sudoku_assay_LeakSw.[1]
- Registry of standard biological parts, Help:Terminators/Measurement/Caitlin Conboy[http://parts.igem.org/Help:Terminators/Measurement/Caitlin_Conboy]
 "
"