Team:NYMU-Taipei/Project/Inhibition/Id-Nosema
From 2013.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
==Introduction== | ==Introduction== | ||
The identification of ''N. apis'' and ''N. ceranae'' is important and still need further research. So far the best differentiation method is the use of species–specific primers; however, the sequence differences are limited to the ribosomal RNA loci, leading to overestimation of Nosema infection rate. Thanks to recent genome study of ''N. apis'' and ''N. ceranae'', we proposed novel primers for differentiation of ''N. spp.'' based on highly conserved, species–specific protein coding genes rather than species–specific sequences in rRNA genes, which is likely to improve the accuracy of species identification. | The identification of ''N. apis'' and ''N. ceranae'' is important and still need further research. So far the best differentiation method is the use of species–specific primers; however, the sequence differences are limited to the ribosomal RNA loci, leading to overestimation of Nosema infection rate. Thanks to recent genome study of ''N. apis'' and ''N. ceranae'', we proposed novel primers for differentiation of ''N. spp.'' based on highly conserved, species–specific protein coding genes rather than species–specific sequences in rRNA genes, which is likely to improve the accuracy of species identification. | ||
| - | |||
| - | |||
==Background== | ==Background== | ||
| + | ===''N. spp.'' Differentiation Methods=== | ||
| + | *Light microscopy method: It is based on morphology difference of ''N. apis'' and ''N. ceranae'', revealing that spores of N. ceranae are oval or rod shaped like a rice, and also slightly thinner than N. apis. Light microscopy method is convenient but not scientific enough since ''N. apis'' bears a striking resemblance to ''N. ceranae''. | ||
| + | === === | ||
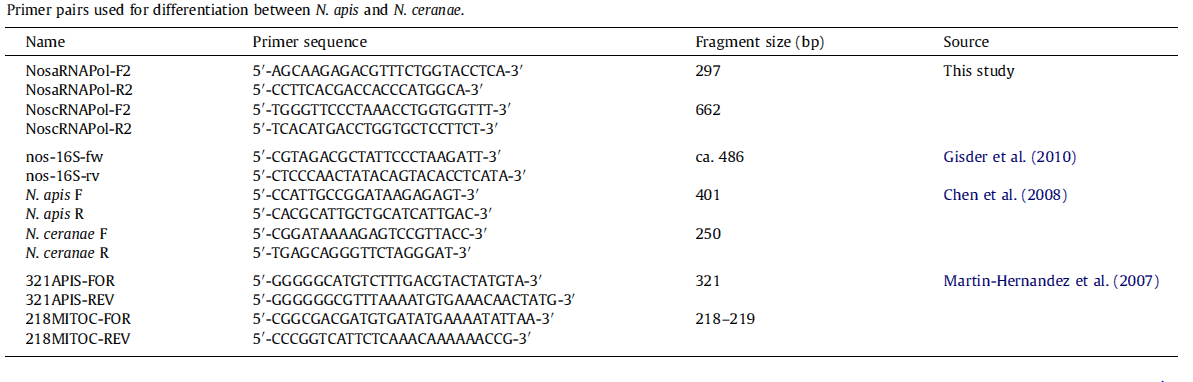
[[File:Nymu-All kinds of primers.PNG|thumb|750px|center|'''Source:Molecular differentiation of Nosema apis and Nosema ceranae based on species–specific sequence differences in a protein coding gene '''<br>http://www.ncbi.nlm.nih.gov/pubmed/23352902]] | [[File:Nymu-All kinds of primers.PNG|thumb|750px|center|'''Source:Molecular differentiation of Nosema apis and Nosema ceranae based on species–specific sequence differences in a protein coding gene '''<br>http://www.ncbi.nlm.nih.gov/pubmed/23352902]] | ||
Revision as of 22:45, 27 September 2013


National Yang Ming University
Contents |
Introduction
The identification of N. apis and N. ceranae is important and still need further research. So far the best differentiation method is the use of species–specific primers; however, the sequence differences are limited to the ribosomal RNA loci, leading to overestimation of Nosema infection rate. Thanks to recent genome study of N. apis and N. ceranae, we proposed novel primers for differentiation of N. spp. based on highly conserved, species–specific protein coding genes rather than species–specific sequences in rRNA genes, which is likely to improve the accuracy of species identification.
Background
N. spp. Differentiation Methods
- Light microscopy method: It is based on morphology difference of N. apis and N. ceranae, revealing that spores of N. ceranae are oval or rod shaped like a rice, and also slightly thinner than N. apis. Light microscopy method is convenient but not scientific enough since N. apis bears a striking resemblance to N. ceranae.
 "
"