Team:Paris Saclay/Project
From 2013.igem.org
EricEscriva (Talk | contribs) (→Detection and degradation of PCB system in Escherichia coli) |
EricEscriva (Talk | contribs) (→References) |
||
| Line 114: | Line 114: | ||
JOURNAL OF BACTERIOLOGY, July 2006 | JOURNAL OF BACTERIOLOGY, July 2006 | ||
| + | |||
| + | |||
Revision as of 17:49, 3 October 2013
Contents |
Detection and degradation of PCB system in Escherichia coli
Since the second half of the XXth century, scientists are fully aware that some bacterial species living in media with high concentration of PCBs are able to degrade PCBs into pyruvate and acetyl-CoA which are then easily metabolized by these organisms.
These bacterial species structure in biofilm with regions that have variable concentrations of oxygen, high at the surface and decreasing with depth. Bacteria living in this habitat have, in most cases, different degradation pathways, which are aerobic or anaerobic depending on their spatial disposition in the biofilm.
Bacteria in aerobic environment use PCB oxidative degradation pathways; those in anaerobic condition degrade PCBs via reductive dechlorination pathways. None of the bacteria seems to use both pathways.
The reductive dechlorination reduces the number of chlorines of high chlorinated PCBs. The dechlorinated PCBs can be further degraded by an oxidative degradation which is efficient only with low chlorinated PCBs. That’s may explain why these different species coexist in biofilms.
Our goal in this project is to desing an organism able to i) detect PCB and then ii)
employ a sequential degradation of the PCB using both combined pathways.
For our experiences, we used bacteria present in nature that are able to detect and degrade the
PCBs, namely [http://en.wikipedia.org/wiki/Burkholderia_xenovorans Burkholderia xenovorans], [http://www.ncbi.nlm.nih.gov/pubmed/22843571Pseudomonas pseudoalcaligenes KF 707] and
[http://en.wikipedia.org/wiki/RhodococcusRhodococcus jostii RHA1].
Construction of a system to detect the presence of PCBs
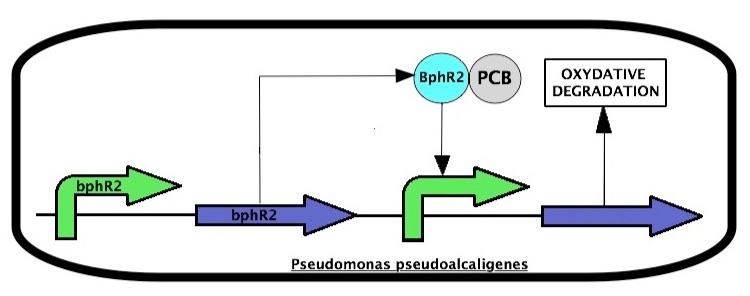
Pseudomonas pseudoalcaligenes expresses enzymes that are responsible for an oxidative degradation of PCBs. The system is regulated two proteins, BphR2 and BphR1 coded by the bphR2 and bphR1 genes, respectively. PCBs induce a BphR2 conformational change to trigger BphR2 transcriptional activity leading to expression of the PCB oxidative degradation pathway.
BphR2 also induces the expression of bphR1.
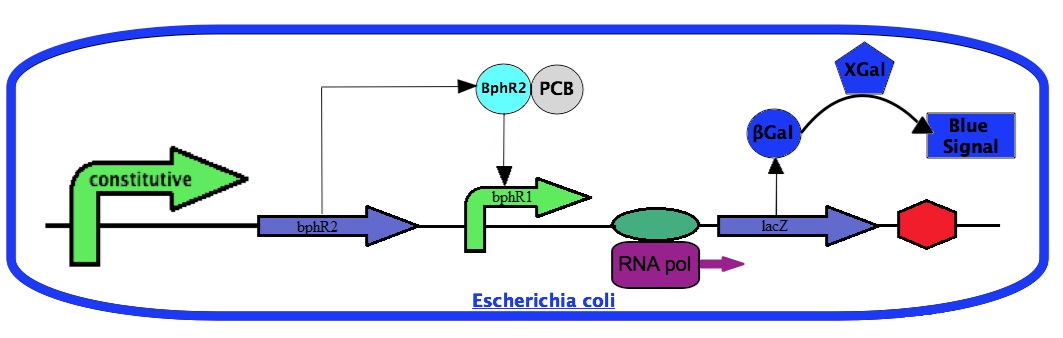
For the project, we will use the bphR2 gene and the bphR1 promoter. We will place the bphR2 coding sequence under a constitutive promoter. We will also construct a transcriptional fusion between the bphR1 promoter with the lacZ gene coding for the β-galactosidase enzyme. The amount of β-galactosidase can be easily monitored by a chemical reaction using Xgal. With this system, the β-galactosidase activity will dependent on the bphR1 promoter expression. Since the bphR1 promoter is controlled by the PCB-activated BphR2, the β-galactosidase activity will correlate with the presence of PCBs.
Combination of the aerobic and anaerobic PCB degradation pathways
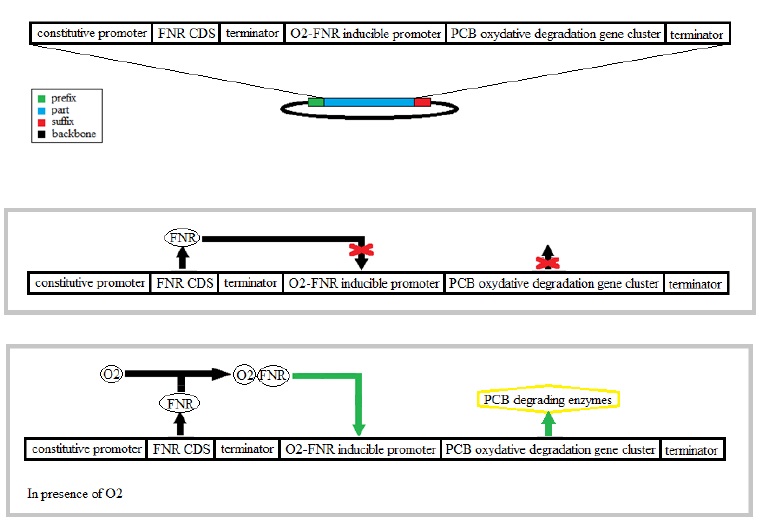
To perform an efficient PCB degradation, two processes should be sequentially combined, the PCB reductive dechlorination followed by a PCB oxidative degradation pathway. Our goal is to engineer a bacterium expressing alternatively both pathways according to growth conditions, with first the reductive dechlorination in anaerobiosis followed by a PCB oxidative degradation in aerobiosis.
The bacterium E. coli has an aerobic and an anaerobic metabolism that will be used to combine of the two PCB degradation pathways. The switch between the aerobic and an anaerobic metabolism is partly regulated the transcriptional regulator FNR. This protein is directly affected by the presence of oxygen which modifies its conformation.
The reductive dechlorination pathway is not well characterized only an enzyme, a dehalogenase, is mentioned as contributing to this pathway. In these anaerobic conditions the chlorine takes the place of the oxygen as the electron acceptor. That’s why we have chosen an activator FNR in presence of oxygen in order to activate the oxidative degradation.
References
Kensuke Furukawa, Hikaru Suenaga and Masatoshi Goto
Biphenyl Dioxygenases: Functional Versatilities and Directed Evolution
JOUNAL OF BACTERIOLOGY, 2004
Kazunari Taira, Jun Hirose, Shinsaku Hayashida, and Kensuke Furukawa
Analysis of bph Operon from the Polychlorynated Biphenyl-degrading Strain of Pseudomonas pseudoalcaligenes KF707
THE JOURNAL OF BIOLOGICAL CHEMISTRY, 1992
Kensuke Furukawa and Hidehiko Fujihara
Microbial Degradation of Polychlorinated Biphenyls: Biochemical and Molecular Features
JOURNAL OF BIOSCIENCE AND BIOENGINEERING, 2008
Jim A. Field, Reyes Sierra-Alvarez
Microbial transformation and degradation of polychlorinated biphenyls
Environmental Pollution, 2008
Lorenz Adrian, Helmut Görisch
Microbial transformation of chlorinated benzenes under anaerobic conditions
Research in Microbiology, 2002
Dean. A Tolla and Michael A. Savageau
Regulation of Aerobic-to-Anaerobic Transitions by the FNR Cycle in Escherichia coli
J. Mol. Biol. (2010)
Hidehiko Fujihara, Hideyuki Yoshida, Tetsuya Matsunaga, Masatoshi Goto, and Kensuke Furukawa
Cross-Regulation of Biphenyl- and Salicylate-Catabolic Genes by Two Regulatory Systems in Pseudomonas pseudoalcaligenes KF707
JOURNAL OF BACTERIOLOGY, July 2006
Article written by Eric
 "
"