Team:Leeds/Results
From 2013.igem.org
m |
m |
||
| Line 74: | Line 74: | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
===Scannning Electron Microscopy=== | ===Scannning Electron Microscopy=== | ||
| - | <gallery caption="SEM Images" widths="225px" heights="220px" perrow="3" mode= | + | <gallery caption="SEM Images" widths="225px" heights="220px" perrow="3" mode=nolines> |
File:SEM Image 1 From Oleg - 1mM TeOS.png | Annotation | File:SEM Image 1 From Oleg - 1mM TeOS.png | Annotation | ||
File:SEM Image 2 From Oleg - 1mM TeOS.png | Annotation | File:SEM Image 2 From Oleg - 1mM TeOS.png | Annotation | ||
Revision as of 14:10, 4 October 2013
Results from the lab so far. Please note, many of the images have been uploaded at high resolution, and are best viewed from their file page. Access this by clicking the image. Different viewing options may be made availalbe, but this is dependent upon Coffee-Induced coding by the Code Monkey.
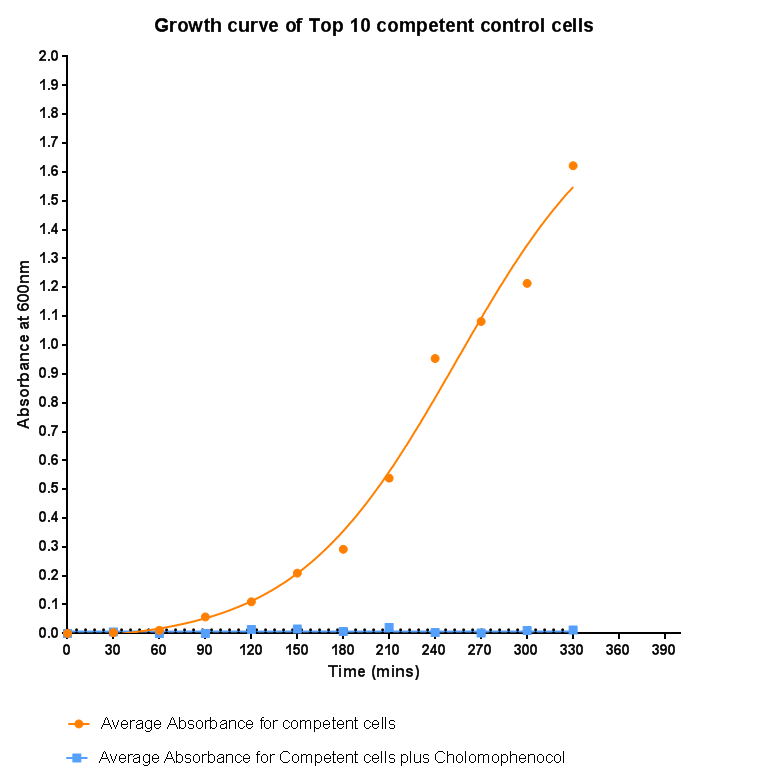
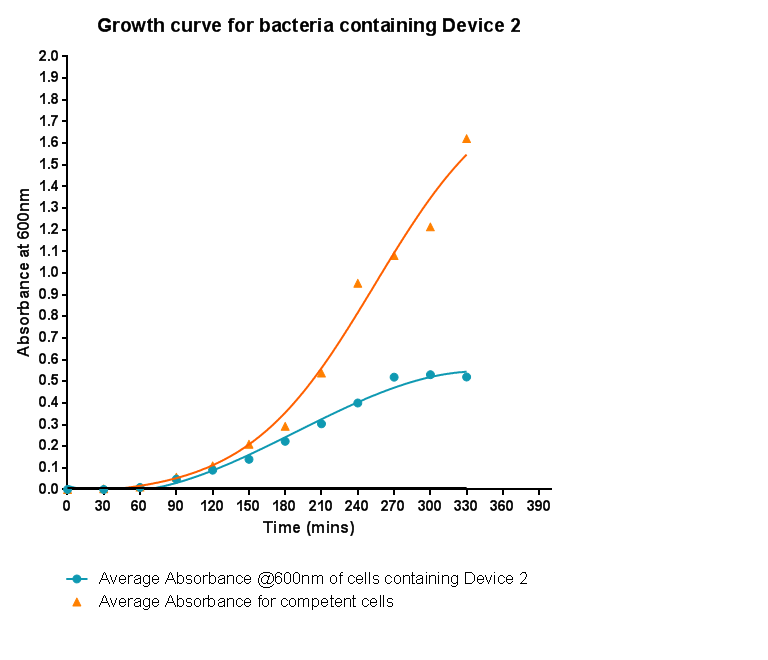
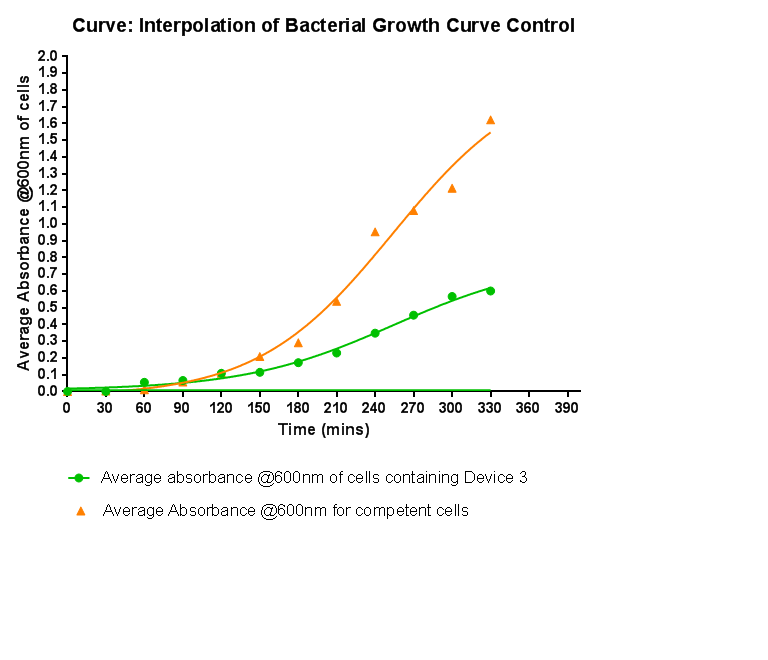
Bacterial growth curvesWe are monitoring the growth of our bacteria containing our devices to see if the genes we inserted have any effect on bacterial growth. Control Bacterial Growth CurveIn this experiment we used our chosen expression cells and monitored their growth on SB media both in the presence of chloramphenicol and without chloramphenicol. As the bacteria do not contain our plasmid that codes for chloramphenicol resistance, we didn't expect them to grow when it was present. The graph produced from the absorbance measurements taken over time, shows the growth of the bacteria. It has a clear lag and log phase when the bacteria are grown in SB media only. But as expected, the graph shows that the bacteria do not grow in the presence of chloramphenicol. This graph will be used as a control and other growth curves of bacteria containing our device will be compared to it in order to determine whether the genes we inserted cause an effect on cell growth.
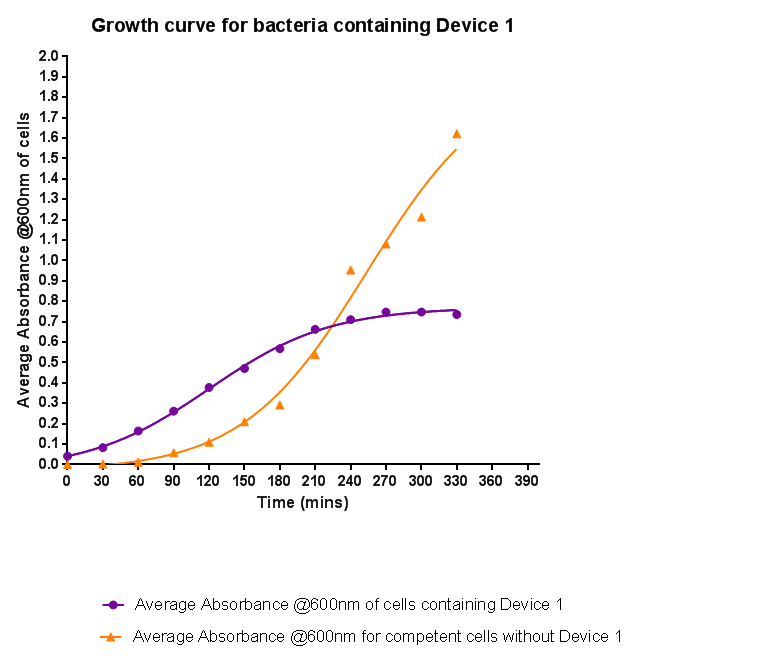
Characterisation of Device 1Growth CurveBacteria were grown which contained Device 1; their absorbance was monitored over time and then the rate of growth compared to bacteria that do not contain Device 1. The graph clearly shows that inserting Device 1 into cells considerably slows their growth in log phase compared with the growth of cells without Device 1 but the lag phase is shorter than in the bacteria without the inserted device. The key difference however is that bacteria containing Device 1 don't grow to as high optical density in log phase as the bacteria without Device 1.
Restriction DigestsText here
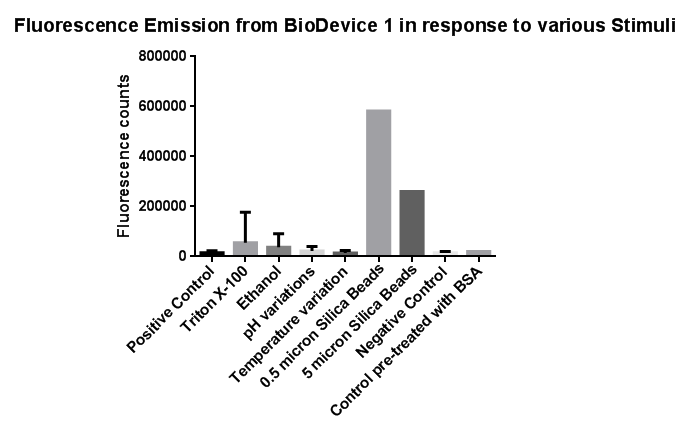
Membrane StressInvestigation of literature regarding the action and regulation of the Cpx pathway lead to the discovery that the system as a whole responds to a large variety of membrane stresses, something that could be true of the specific promoter we chose to use (pCpxR). As a result of this we deemed it necessary to investigate the response of our promoter to each individual membrane stress.
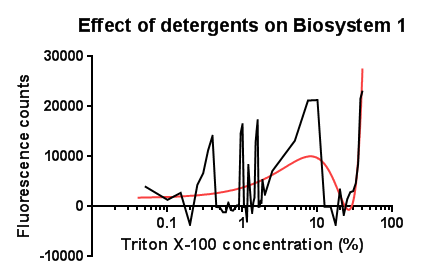
Triton X-100<p align="justify">Between the concentration values of 0.1-9% triton X-100 the data shows roughly the relationship that was expected, a rise in GFP production as the triton concentration increases. This is likely due to triton acting as a detergent resulting in damage to the plasma membrane of cells, activating the Cpx pathway in general, including the cpxR promoter. Past 10% Triton X-100 concentration it is assumed that cells are dying in large quantities, being unable to produce GFP at all or in very limited amounts before they are killed. This explains the corrected fluorescence readings reducing to below zero.
pH EffectsThe slight fluctuations across the pH scale are in line with the background fluorescence readings from our cell cultures, representing the standard levels of cpx activation as the cells collide, disperse in solution and divide during replication.
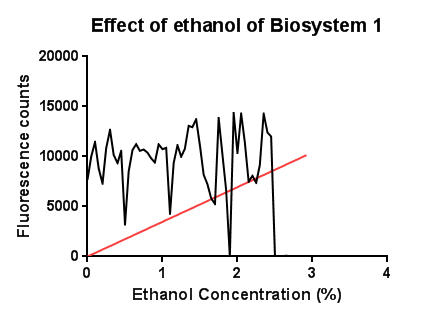
EthanolIn this test a large, seemingly anomalous spike in an initial experiment was re-tested with smaller increasing increments of ethanol. The results are somewhat erratic but the peak just before 3% ethanol can be seen, before the fluorescence reading goes on to trough again.
Membrane Stress- Atomic Force MicroscopyAtomic Force Microscopy utilises Van-Der-Waals forces to attract the tip towards a surface, causing the bending of a cantilever, onto which a laser is pointed. The tip deflection causes the laser light to also be deflected and this is tracked using a bank of photodiodes. AFM is typically used to image surfaces at the atomic scale, as well as for Force Spectroscopy. When used in Force Spectroscopy, the tip is brought into direct contact with the sample, allowing the sample protein to bind to the silicon tip. Then, the AFM is slowly retracted at varying forces or to varying distances, applying a force to the protein. This causes portions of the protein to unravel, and the distinct patterns associated with α-helices and β-pleated sheets can be seen in this data. This allows the exact secondary and tertiary structure of a protein to be determined, but also the energy profile of various conformations, which in turn tells us how the protetin folds.
Characterisation of Si4Growth CurveHere the growth of cells containing Si4 was monitored via absorbance and compared to the growth of cells that do not contain Si4. As was seen when device 1 was inserted into the cells, the growth is slowed down considerably. Although the graph shows a log and a lag phase for the cells containing Si4, the absorbance reached in log phase is very low compared to cells without Si4; it is even lower than the absorbance reached for cells containing Device 1.
Restriction Digeststext here, do not edit outside of the tags
Characterisation of Device 3Growth CurveThis graph shows the results of monitoring the growth of cells with contain Device 3. As with cells containing Device 1 and Device 2, the growth of the cells is considerably slowed down when compared with cells that do not contain any device at all. A slight change in the gradient of the graph can be seen showing the transition of the cell growth from lag to log phase, so the cells are still growing normally, but the insertion of Device 3 is restricting the growth of the cells.
Restriction Digeststext here, do not edit outside of the tags
Confocal Microscopytext here, do not edit outside of the tags
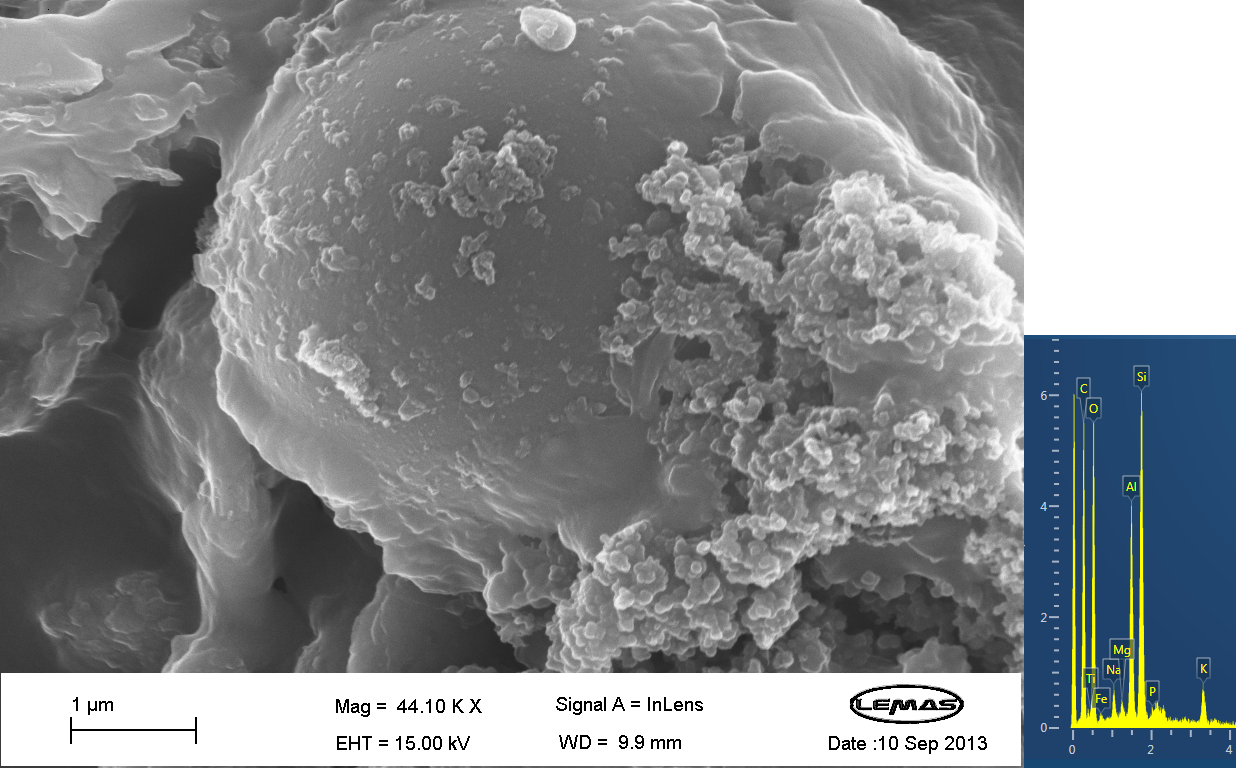
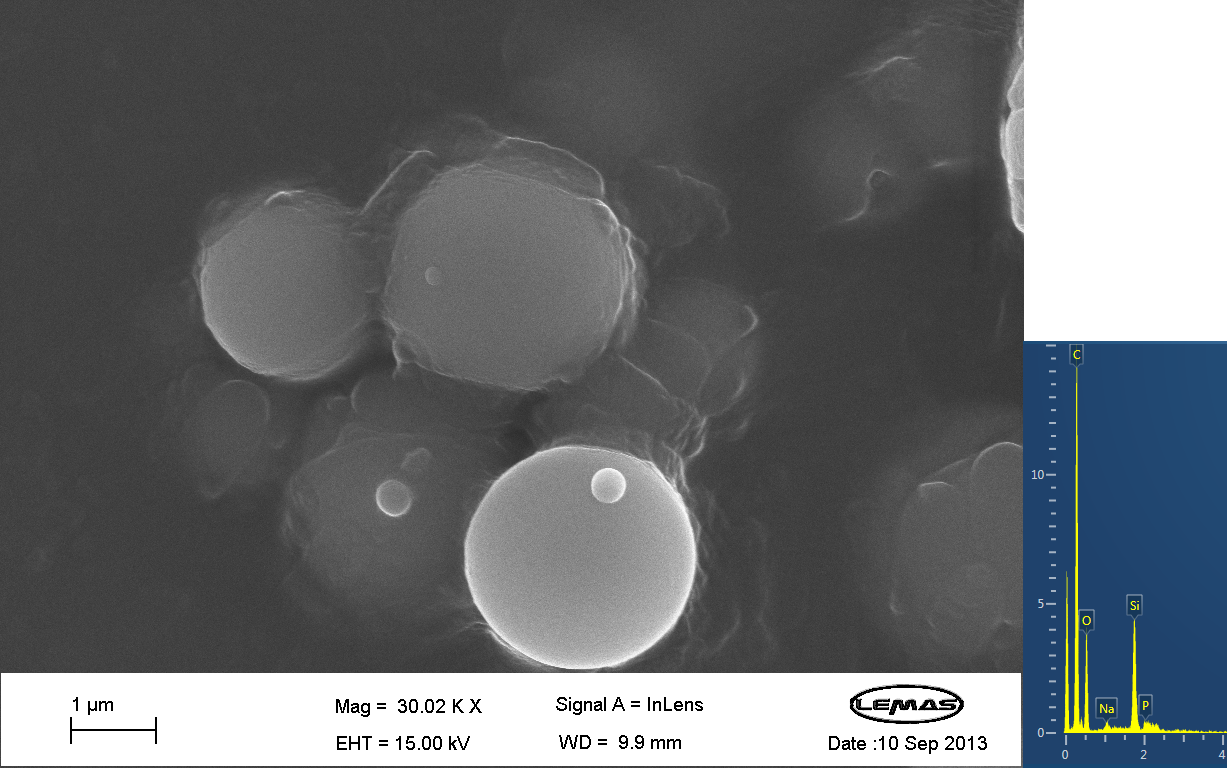
Scannning Electron MicroscopyThe gallery above contains images that were produced by the Scanning Electron Microscope (SEM) at the University of Leeds. Device 2 was incubated in solution of varying concentrations of a silicate compound, which was solubilized using the sol-gel method from Tetraethyl orthosilicate (TEOS). Samples were incubated at room temperature, as well as some were done at 45ºC. For the control samples, cells without the Device 2 construct gene were used, and did not react with the silicate compound.
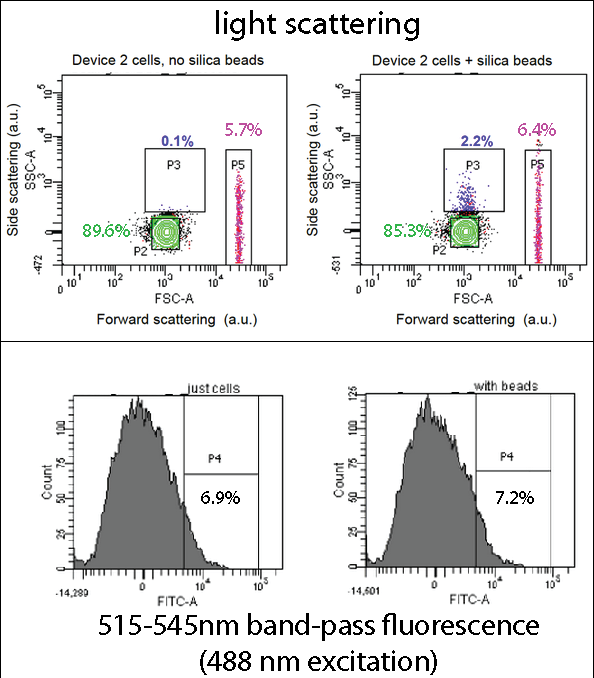
FACSDevice 2 contains our INP+Si4 construct, along with the Cpx and GFP genes. It was characterized using a flow cytometry method called Fluorescence-activated cell sorting (FACS) in solution with and without beads. The FASC images contain Side scattering and Forward scattering information, relating to relative size and density of the cells.
| ||||||||
 |
| |||||||

| ||||||||

| ||||||||
 "
"