Team:Paris Saclay/Notebook/July/17
From 2013.igem.org
(→1 - Electrophoresis of digestions of BBa_B0015, BBa_B0017, BBa_I732019 by EcoRI/PstI) |
|||
| (18 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Paris_Saclay/incl_debut_generique}} | {{Team:Paris_Saclay/incl_debut_generique}} | ||
| - | ='''Notebook : July | + | ='''Notebook : July 17'''= |
=='''Lab work'''== | =='''Lab work'''== | ||
| Line 7: | Line 7: | ||
==='''A - Aerobic/Anaerobic regulation system'''=== | ==='''A - Aerobic/Anaerobic regulation system'''=== | ||
| - | ===='''Objective : obtaining | + | ===='''Objective : obtaining BBa_K1155003, BBa_K1155007'''==== |
| - | ===='''1 - | + | ===='''1 - Electrophoresis of digestions of BBa_B0015, BBa_B0017, BBa_I732019 by EcoRI/PstI'''==== |
| - | + | Anaïs, Sheng | |
| - | + | ||
| - | + | ||
| + | * BBa_B0015, BBa_B0017 : | ||
| + | {| | ||
| + | | style="width:350px;border:1px solid black;" |[[File:Psgel11707.jpg|300px]] | ||
| + | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| + | * Well 1 : 2µL BBa_B0015+1µL of 6X loading dye | ||
| + | * Well 2 : 2µL BBa_B0015+1µL of 6X loading dye | ||
| + | * Well 3 : 2µL BBa_B0015+1µL of 6X loading dye | ||
| + | * Well 4 : 6µL DNA Ladder | ||
| + | * Well 5 : 2µL BBa_B0017+1µL of 6X loading dye | ||
| + | * Well 6 : 2µL BBa_B0017+1µL of 6X loading dye | ||
| + | * Well 7 : 2µL BBa_B0017+1µL of 6X loading dye | ||
| + | * Gel : 1% | ||
| + | |} | ||
| + | Expected sizes : | ||
| + | * BBa_B0015 digested by EcoRI/PstI : 500bp | ||
| + | * BBa_B0017 digested by EcoRI/PstI : 500bp | ||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We obtain fragments at the right size. We will do a PCR of them. | ||
| + | |} | ||
| + | {| | ||
| + | | style="width:350px;border:1px solid black;" |[[File:Psgel31707.jpg|300px]] | ||
| + | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| + | * Well 1 : 5µL BBa_I732019 digested by EcoRI/PstI+1µL of 6X loading dye | ||
| + | * Well 2 : 6µL DNA Ladder | ||
| + | * Gel : 1% | ||
| + | |} | ||
| + | Expected sizes : | ||
| + | * BBa_I732019 : 3230bp | ||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We didn't obtain fragments at the right size. We will use BBa_I732017. | ||
| + | |} | ||
| + | ==='''B - PCB sensor system'''=== | ||
| + | ===='''Objective : obtaining BBa_K1155001, BBa_K1155002 and BphR2'''==== | ||
| + | ===='''1 - Stock of BBa_K1155001'''==== | ||
| + | Anaïs, Sheng | ||
| + | Used quantities : | ||
| + | * BBa_K1155001 confirmed : 1 mL | ||
| + | * Glycerol : 500µL glycerol. | ||
| + | We stocked them at -20°C. | ||
| + | ===='''2 - Extraction of BBa_K1155001 from DH5α'''==== | ||
| + | Anaïs, Sheng | ||
| + | Protocol : [[Team:Paris_Saclay/extraction|Hight copy plamid extraction]] | ||
=='''Modeling'''== | =='''Modeling'''== | ||
| Line 58: | Line 100: | ||
Ethic about open source:<br> | Ethic about open source:<br> | ||
-Hsim is not an open source software, it belongs to the University of Paris Sud. However, it is a free software, we can download it free.<br> | -Hsim is not an open source software, it belongs to the University of Paris Sud. However, it is a free software, we can download it free.<br> | ||
| - | |||
<br> | <br> | ||
| Line 67: | Line 108: | ||
|[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | |[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | ||
| - | |[[Team:Paris Saclay/Notebook/July/18|<big> | + | |[[Team:Paris Saclay/Notebook/July/18|<big>Next day</big>]] |
|} | |} | ||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Latest revision as of 00:11, 5 October 2013
Contents |
Notebook : July 17
Lab work
A - Aerobic/Anaerobic regulation system
Objective : obtaining BBa_K1155003, BBa_K1155007
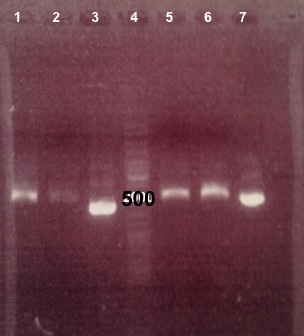
1 - Electrophoresis of digestions of BBa_B0015, BBa_B0017, BBa_I732019 by EcoRI/PstI
Anaïs, Sheng
- BBa_B0015, BBa_B0017 :
Expected sizes :
- BBa_B0015 digested by EcoRI/PstI : 500bp
- BBa_B0017 digested by EcoRI/PstI : 500bp
|
We obtain fragments at the right size. We will do a PCR of them. |
|
|
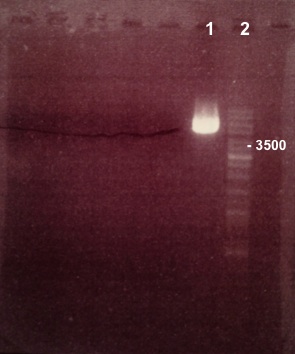
Expected sizes :
- BBa_I732019 : 3230bp
|
We didn't obtain fragments at the right size. We will use BBa_I732017. |
B - PCB sensor system
Objective : obtaining BBa_K1155001, BBa_K1155002 and BphR2
1 - Stock of BBa_K1155001
Anaïs, Sheng
Used quantities :
- BBa_K1155001 confirmed : 1 mL
- Glycerol : 500µL glycerol.
We stocked them at -20°C.
2 - Extraction of BBa_K1155001 from DH5α
Anaïs, Sheng
Protocol : Hight copy plamid extraction
Modeling
Meeting memo
Meeting Date: 17/07/2013
Meeting participants: Patrick Amar, Claire Toffano-Nioche, Abdou Mouhtare, Damir Damipator, Gabriel Gallin, Zhou Yi
Recorder: Zhou Yi
Hsim:
-Hsim is created for bio-chemical reaction simulation, it demonstrates bio-chemical systems which the scientists are already known.
-The molecules are simulated as spheres (in 2D or 3D). Initial positions and size of sphere are configurable.
-example of a Hsim command:
B + V -> R + V [probability]
-Molecules diffusion are simulated by movements of spheres, if the distance between two centers of the spheres is less than the sum of 2 radium, we consider that will lead to a collision.
-Computer takes a random sample and compare it to the probability which is specified in equation. If the value exceeds the threshold, we consider that the reaction takes place.
-Probability of success is associated with kinetics of reaction which depends on concentrations of molecules varying during the reaction.
-Time unit is 100ms. So movement of spheres is discreet.
-Spheres move randomly but could follow some stochastic rules (like Brownian movement).
Ethic about open source:
-Hsim is not an open source software, it belongs to the University of Paris Sud. However, it is a free software, we can download it free.
| Previous week | Back to calendar | Next day |
 "
"