Team:Paris Saclay/Notebook/August/13
From 2013.igem.org
(→2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI) |
|||
| (67 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
==='''A - Aerobic/Anaerobic regulation system'''=== | ==='''A - Aerobic/Anaerobic regulation system'''=== | ||
| - | ===='''Objective : | + | ===='''Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006'''==== |
| - | ===='''1 - Electrophoresis to check the digestion of | + | ===='''1 - Electrophoresis to check the digestion of BBa_K1155000 by SpeI/PstI'''==== |
Nadia | Nadia | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel11308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 5µL | + | * Well 2 : 5µL BBa_K1155000 digested by SpeI/PstI +1µl of 6X loading dye |
* Gel : 0.8% | * Gel : 0.8% | ||
|} | |} | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | * Pndh* : 111bp |
| - | * | + | * pSB1C3 : 2070bp |
| - | + | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We can't see any band for | + | We can't see any band for BBa_K1155000 digestion. The digestion failed because we used the wrong enzymes. We will do it again with the good ones. |
|} | |} | ||
| - | ====2 - Digestion of | + | ===='''2 - Extraction of BBa_K1155004, BBa_1155005, BBa_K1155006 from DH5αstrain'''==== |
| + | |||
| + | XiaoJing | ||
| + | |||
| + | Protocol : [[Team:Paris_Saclay/extraction|High-copy plasmid extraction]] | ||
| + | |||
| + | ===='''3 - Digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI'''==== | ||
Anaïs, Nadia | Anaïs, Nadia | ||
| - | * | + | Used quantities : |
| + | |||
| + | * BBa_K1155000 : | ||
** Buffer FD : 5µL | ** Buffer FD : 5µL | ||
** H2O : 39µL | ** H2O : 39µL | ||
| Line 40: | Line 47: | ||
** SpeI FD : 1µL | ** SpeI FD : 1µL | ||
| - | * | + | * BBa_K1155000 : |
** Buffer FD : 5µL | ** Buffer FD : 5µL | ||
** H2O : 38µL | ** H2O : 38µL | ||
| Line 47: | Line 54: | ||
**EcoRI FD : 1µL | **EcoRI FD : 1µL | ||
| - | * | + | * BBa_K1155004, BBa_K1155005, BBa_K1155006 : |
** Buffer FD : 2µL | ** Buffer FD : 2µL | ||
** H2O : 7µL | ** H2O : 7µL | ||
| Line 53: | Line 60: | ||
** SpeI FD : 1µL | ** SpeI FD : 1µL | ||
| - | * | + | * BBa_K1155004, BBa_K1155005, BBa_K1155006 : |
** Buffer FD : 2µL | ** Buffer FD : 2µL | ||
** H2O : 6µL | ** H2O : 6µL | ||
| Line 60: | Line 67: | ||
** EcoRI FD : 1µL | ** EcoRI FD : 1µL | ||
| - | ==== | + | We incubat the digestion at 37°C during 10 minutes. |
| + | |||
| + | ====4 - '''Electrophoresis to check the digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI'''==== | ||
Anaïs, Nadia | Anaïs, Nadia | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel21308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 5µL | + | * Well 2 : 5µL BBa_K1155000 digested by SpeI + 1µl of 6X loading dye |
| - | * Well 3 : 5µL | + | * Well 3 : 5µL BBa_K1155000 +1µl of 6X loading dye |
| - | * Well 4 : 5µL | + | * Well 4 : 5µL BBa_K1155000 digested by EcoRI/Spe I + 1µl of 6X loading dye |
* Gel : 0.8% | * Gel : 0.8% | ||
|} | |} | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | * Pndh* : 111bp |
| - | * | + | * pSB1C3 : 2070 kb |
| - | + | ||
| - | + | ||
{| | {| | ||
| Line 86: | Line 93: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel31308.jpg|500px]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
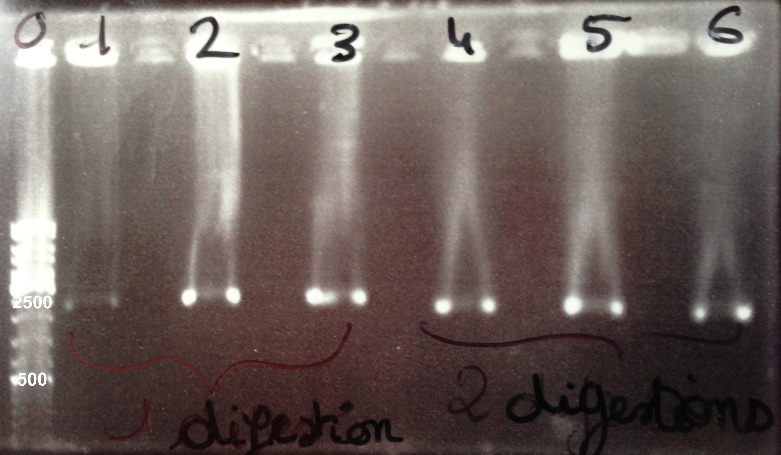
| - | * Well | + | * Well 0 : 6µL DNA Ladder |
| - | * Well | + | * Well 1 : 5µL BBa_K1155006 digested by SpeI + 1µl of 6X loading dye |
| - | * Well | + | * Well 2 : 5µL BBa_K1155005 digested by SpeI + 1µl of 6X loading dye |
| - | * Well | + | * Well 3 : 5µL BBa_K1155004 digested by SpeI + 1µl of 6X loading dye |
| - | * Well | + | * Well 4 : 5µL BBa_K1155006 digested by EcoRI/SpeI + 1µl of 6X loading dye |
| - | * Well | + | * Well 5 : 5µL BBa_K1155005 digested by EcoRI/SpeI + 1µl of 6X loading dye |
| - | * Well | + | * Well 6 : 5µL BBa_K1155004 digested by EcoRI/SpeI + 1µl of 6X loading dye |
* Gel : 0.8% | * Gel : 0.8% | ||
|} | |} | ||
| Line 100: | Line 107: | ||
Expected sizes : | Expected sizes : | ||
* NarK, Nar G, NirB : 200kb | * NarK, Nar G, NirB : 200kb | ||
| - | * | + | * pSB1C3 : 2070bp |
| - | * NarK in | + | * NarK in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We lost all our digestion product so we will do it again | + | We lost all our double digestion product so we will do it again. We obtain BBa_K1155004, BBa_K1155005, BBa_K1155006 digested by SpeI fragments at the right size. We can purify it. |
|} | |} | ||
| - | ==== | + | ===='''5 - Liquid culture of MG1655Z1 Δfnr::Km'''==== |
| + | |||
| + | XiaoJing | ||
| + | |||
| + | We picked up one colony in 5mL of LB and 5µL of kanamycine. We do it twice. | ||
| + | |||
| + | We incubated our culture at 37°C with agigation at 150rpm. | ||
| + | |||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We obtain fragments at the right size. | ||
| + | |} | ||
| + | |||
| + | ===='''Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3'''==== | ||
| + | |||
| + | ===='''1 - Digestion of BBa_J04450 by EcoRI/PstI'''==== | ||
Anaïs | Anaïs | ||
| + | |||
| + | Used quantities : | ||
* Buffer FD: 2µL | * Buffer FD: 2µL | ||
| Line 118: | Line 142: | ||
* PstI FD : 1µL | * PstI FD : 1µL | ||
| - | ==== | + | We incubated the digestion at 37°C during 10 minutes. |
| + | |||
| + | ===='''2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI'''==== | ||
Anaïs, Nadia | Anaïs, Nadia | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel41308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 4 : 6µL DNA Ladder | * Well 4 : 6µL DNA Ladder | ||
| - | * Well 7 : 5µL of | + | * Well 7 : 5µL of BBa_J04450 digested by EcoRI/PstI + 1µL of 6X loading dye |
* Gel : 0.8% | * Gel : 0.8% | ||
|} | |} | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | * pSB3K3 : 2750bp |
| - | * GFP : | + | * GFP : 1069bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | + | We obtained a fragment at the right size. | |
|} | |} | ||
| - | |||
==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ||
| - | ===='''Objective : obtaining FNR and BphR2 proteins'''==== | + | ===='''Objective : obtaining FNR and BphR2 proteins (Gibson assembly)'''==== |
===='''1 - PCR of FRN Part I, FNR Part II, BphR2 Part I'''==== | ===='''1 - PCR of FRN Part I, FNR Part II, BphR2 Part I'''==== | ||
| Line 148: | Line 173: | ||
Anaïs | Anaïs | ||
| - | + | Used quantities : | |
| + | * Bphr2 Part I : | ||
| + | ** Oligo 54F : 1µL | ||
| + | ** Oligo 55R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA of ''Pseudomonas pseudoalcaligenes'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * FNR Part I : | ||
| + | ** Oligo 59F : 1µL | ||
| + | ** Oligo 60R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Escherichia coli'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * FNR Part II : | ||
| + | ** Oligo 61F : 1µL | ||
| + | ** Oligo 62R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Escherichia coli'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | PCR Program : | ||
| + | |||
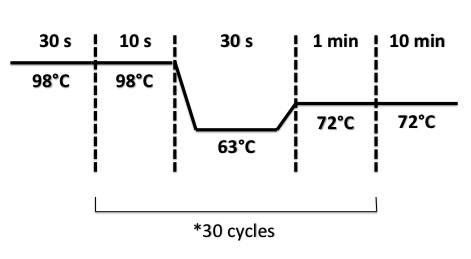
| + | * BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I : | ||
| + | |||
| + | [[File:PsPCRBphR23007.jpg|400px]] | ||
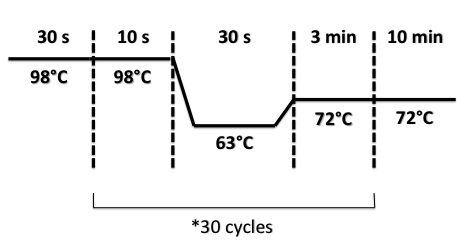
| + | |||
| + | * FNR Part I, FNR Part II, RBS-FNR Part I : | ||
| + | |||
| + | [[File:PsPCRFNR3007.jpg|400px]] | ||
===='''2 - Electrophoresis of PCR products : FRN Part I, FNR Part II, BphR2 Part I'''==== | ===='''2 - Electrophoresis of PCR products : FRN Part I, FNR Part II, BphR2 Part I'''==== | ||
| Line 155: | Line 216: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel51308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| - | * Well 1 : 5µL FNR Part II+1µl of 6X loading dye | + | * Well 1 : 5µL FNR Part II + 1µl of 6X loading dye |
| - | * Well 2 : 5µL BphR2 Part I+1µl of 6X loading dye | + | * Well 2 : 5µL BphR2 Part I + 1µl of 6X loading dye |
| - | * Well 3 : 5µL FNR Part I+1µl of 6X loading dye | + | * Well 3 : 5µL FNR Part I + 1µl of 6X loading dye |
* Well 4 : 6µL DNA Ladder | * Well 4 : 6µL DNA Ladder | ||
* Gel : 1% | * Gel : 1% | ||
| Line 165: | Line 226: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel61308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| - | * Well 1 : 5µL FNR Part II+1µl of 6X loading dye | + | * Well 1 : 5µL FNR Part II + 1µl of 6X loading dye |
| - | * Well 2 : 5µL BphR2 Part I+1µl of 6X loading dye | + | * Well 2 : 5µL BphR2 Part I + 1µl of 6X loading dye |
| - | * Well 3 : 5µL FNR Part I+1µl of 6X loading dye | + | * Well 3 : 5µL FNR Part I + 1µl of 6X loading dye |
* Well 4 : 6µL DNA Ladder | * Well 4 : 6µL DNA Ladder | ||
* Gel : 1% | * Gel : 1% | ||
| Line 175: | Line 236: | ||
Expected sizes : | Expected sizes : | ||
| - | * FNR | + | * FNR Part I : 597 kb |
| - | * FNR | + | * FNR Part II : 200 kb |
| - | * BphR2 | + | * BphR2 Part I : 178 kb |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
It's impossible to read the first gel. We do it again. | It's impossible to read the first gel. We do it again. | ||
| - | In the second gel, we obtain FNR Part I and FNR Part II fragments at the right size. We can purify it. We also obtain BphR2 frangment at the right size but it was mix with other DNA frangments. We will try to make a gel purification of it. | + | In the second gel, we obtain FNR Part I and FNR Part II fragments at the right size. We can purify it. We also obtain BphR2 Part I frangment at the right size but it was mix with other DNA frangments. We will try to make a gel purification of it. |
|} | |} | ||
| - | ===='''3 - | + | ===='''3 - Electrophoresis of PCR product : BphR2 Part I'''==== |
Anaïs, Damir, Nadia | Anaïs, Damir, Nadia | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" |[[]] | + | | style="width:350px;border:1px solid black;" |[[File:Psgel71308.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 40µL of BphR2 Part I+8µL of 6X loading dye | + | * Well 2 : 40µL of BphR2 Part I + 8µL of 6X loading dye |
* Gel : 1% | * Gel : 1% | ||
|} | |} | ||
| - | Protocol : [ | + | Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ] |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | Even if the two lightest stripes are overlapping, aims to do a second gel | + | Even if the two lightest stripes are overlapping, aims to do a second gel purification with these two stripes, we did the gel purification . After argumentation, we decided to do the PCR of BphR2 Part I again thanks to new PCR program and new quantities. |
|} | |} | ||
| Line 208: | Line 269: | ||
Anaïs | Anaïs | ||
| - | + | Used quantities : | |
| - | + | ||
| - | + | ||
* Oligo 54F : 1µL | * Oligo 54F : 1µL | ||
* Oligo 55R : 1µL | * Oligo 55R : 1µL | ||
| Line 218: | Line 277: | ||
* Phusion : 1µL | * Phusion : 1µL | ||
* H2O : 33.5µL | * H2O : 33.5µL | ||
| + | |||
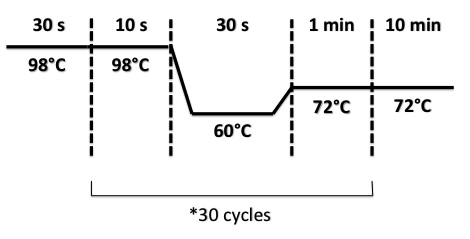
| + | PCR Program : | ||
| + | |||
| + | [[File:PsPCRBphR2PI1308.jpg|400px]] | ||
| + | |||
| + | '''5 - Gel purification of PCR product : FNR Part I, FNR Part II, RBS-FNR Part I, BphR2 Part II, | ||
| + | '''RBS-BphR2 Part I'''''' | ||
| + | |||
| + | XiaoJing | ||
| + | |||
| + | Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ] | ||
| + | |||
| + | Nanodrop : | ||
| + | * FNR Part I : 152.7ng/µL | ||
| + | * FNR Part II : 137.2ng/µL | ||
| + | * RBS-FNR Part I : 153.7ng/µL | ||
| + | * BphR2 Part II : 129.9ng/µL | ||
| + | * RBS-BphR2 Part I : 158.1ng/µL | ||
| + | |||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | The purification was good. We will do the Gibson assembly. | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| border="1" align="center" | ||
| + | |[[Team:Paris Saclay/Notebook/August/12|<big>Previous day</big>]] | ||
| + | |||
| + | |[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | ||
| + | |||
| + | |[[Team:Paris Saclay/Notebook/August/14|<big>Next day</big>]] | ||
| + | |} | ||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Latest revision as of 01:28, 5 October 2013
Notebook : August 13
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
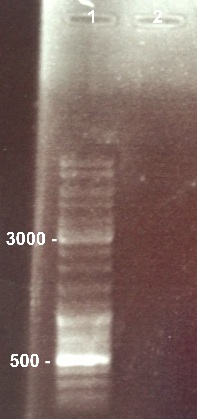
1 - Electrophoresis to check the digestion of BBa_K1155000 by SpeI/PstI
Nadia
|
|
Expected sizes :
- Pndh* : 111bp
- pSB1C3 : 2070bp
|
We can't see any band for BBa_K1155000 digestion. The digestion failed because we used the wrong enzymes. We will do it again with the good ones. |
2 - Extraction of BBa_K1155004, BBa_1155005, BBa_K1155006 from DH5αstrain
XiaoJing
Protocol : High-copy plasmid extraction
3 - Digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI
Anaïs, Nadia
Used quantities :
- BBa_K1155000 :
- Buffer FD : 5µL
- H2O : 39µL
- DNA : 5µL
- SpeI FD : 1µL
- BBa_K1155000 :
- Buffer FD : 5µL
- H2O : 38µL
- DNA : 5µL
- SpeI FD : 1µL
- EcoRI FD : 1µL
- BBa_K1155004, BBa_K1155005, BBa_K1155006 :
- Buffer FD : 2µL
- H2O : 7µL
- DNA : 10µL
- SpeI FD : 1µL
- BBa_K1155004, BBa_K1155005, BBa_K1155006 :
- Buffer FD : 2µL
- H2O : 6µL
- DNA : 10µL
- SpeI FD : 1µL
- EcoRI FD : 1µL
We incubat the digestion at 37°C during 10 minutes.
4 - Electrophoresis to check the digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and by SpeI/EcoRI
Anaïs, Nadia
Expected sizes :
- Pndh* : 111bp
- pSB1C3 : 2070 kb
|
We lost all our digestion product so we will do it again. |
Expected sizes :
- NarK, Nar G, NirB : 200kb
- pSB1C3 : 2070bp
- NarK in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp
|
We lost all our double digestion product so we will do it again. We obtain BBa_K1155004, BBa_K1155005, BBa_K1155006 digested by SpeI fragments at the right size. We can purify it. |
5 - Liquid culture of MG1655Z1 Δfnr::Km
XiaoJing
We picked up one colony in 5mL of LB and 5µL of kanamycine. We do it twice.
We incubated our culture at 37°C with agigation at 150rpm.
|
We obtain fragments at the right size. |
Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3
1 - Digestion of BBa_J04450 by EcoRI/PstI
Anaïs
Used quantities :
- Buffer FD: 2µL
- H2O : 6µL
- DNA : 10µL
- EcoRI FD : 1µL
- PstI FD : 1µL
We incubated the digestion at 37°C during 10 minutes.
2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI
Anaïs, Nadia

|
|
Expected sizes :
- pSB3K3 : 2750bp
- GFP : 1069bp
|
We obtained a fragment at the right size. |
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR and BphR2 proteins (Gibson assembly)
1 - PCR of FRN Part I, FNR Part II, BphR2 Part I
Anaïs
Used quantities :
- Bphr2 Part I :
- Oligo 54F : 1µL
- Oligo 55R : 1µL
- Buffer Phusion : 10µL
- DNA of Pseudomonas pseudoalcaligenes : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part I :
- Oligo 59F : 1µL
- Oligo 60R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part II :
- Oligo 61F : 1µL
- Oligo 62R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
PCR Program :
- BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I :
- FNR Part I, FNR Part II, RBS-FNR Part I :
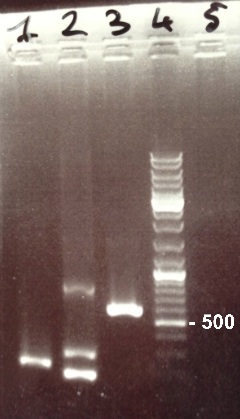
2 - Electrophoresis of PCR products : FRN Part I, FNR Part II, BphR2 Part I
Damir

|
|
|
|
Expected sizes :
- FNR Part I : 597 kb
- FNR Part II : 200 kb
- BphR2 Part I : 178 kb
|
It's impossible to read the first gel. We do it again. In the second gel, we obtain FNR Part I and FNR Part II fragments at the right size. We can purify it. We also obtain BphR2 Part I frangment at the right size but it was mix with other DNA frangments. We will try to make a gel purification of it. |
3 - Electrophoresis of PCR product : BphR2 Part I
Anaïs, Damir, Nadia

|
|
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
|
Even if the two lightest stripes are overlapping, aims to do a second gel purification with these two stripes, we did the gel purification . After argumentation, we decided to do the PCR of BphR2 Part I again thanks to new PCR program and new quantities. |
4 - PCR of BphR2 Part I
Anaïs
Used quantities :
- Oligo 54F : 1µL
- Oligo 55R : 1µL
- DNA : 2µL
- Buffer Phusion : 10µL
- dNTP : 1µL
- Phusion : 1µL
- H2O : 33.5µL
PCR Program :
5 - Gel purification of PCR product : FNR Part I, FNR Part II, RBS-FNR Part I, BphR2 Part II, RBS-BphR2 Part I'
XiaoJing
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
Nanodrop :
- FNR Part I : 152.7ng/µL
- FNR Part II : 137.2ng/µL
- RBS-FNR Part I : 153.7ng/µL
- BphR2 Part II : 129.9ng/µL
- RBS-BphR2 Part I : 158.1ng/µL
|
The purification was good. We will do the Gibson assembly. |
| Previous day | Back to calendar | Next day |
 "
"