Team:Leeds/Modeling
From 2013.igem.org
m |
|||
| (17 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
</html> | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| - | {{Team:Leeds_layout|Header=Modelling|content= | + | {{Team:Leeds_layout|HeaderParky=</div>[[File:Leeds_Parkytopmodel.png|150px|link=|frameless]]<div>|HeaderImage=[[File:Leeds_Modelheader.png|link=|825px|frameless]]|Header=Modelling|content= |
We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project. | We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project. | ||
<br> | <br> | ||
| Line 17: | Line 17: | ||
We are currently using [http://bionetgen.org/index.php/Main_Page BioNetGenLanguage] to write the model code, as this provides a rule-based coding language that is easier to use than SBML. Rule-based modelling allows mechanistic simulation of the pertinent reactants, reducing the need to know the exact kinetics of reactions, as sub-processes can be absorbed into rate constants. Additionally, adaptation of the model should be easier after a basic framework has been produced, as new processes simply require insertion of new rules into the code. Finally, BNGL offers easy integration with [http://www.ibiblio.org/virtualcell/ Virtual Cell], a graphical simulator of cell systems - ideal for outreach with the public and teaching. | We are currently using [http://bionetgen.org/index.php/Main_Page BioNetGenLanguage] to write the model code, as this provides a rule-based coding language that is easier to use than SBML. Rule-based modelling allows mechanistic simulation of the pertinent reactants, reducing the need to know the exact kinetics of reactions, as sub-processes can be absorbed into rate constants. Additionally, adaptation of the model should be easier after a basic framework has been produced, as new processes simply require insertion of new rules into the code. Finally, BNGL offers easy integration with [http://www.ibiblio.org/virtualcell/ Virtual Cell], a graphical simulator of cell systems - ideal for outreach with the public and teaching. | ||
===Cpx Pathway Model=== | ===Cpx Pathway Model=== | ||
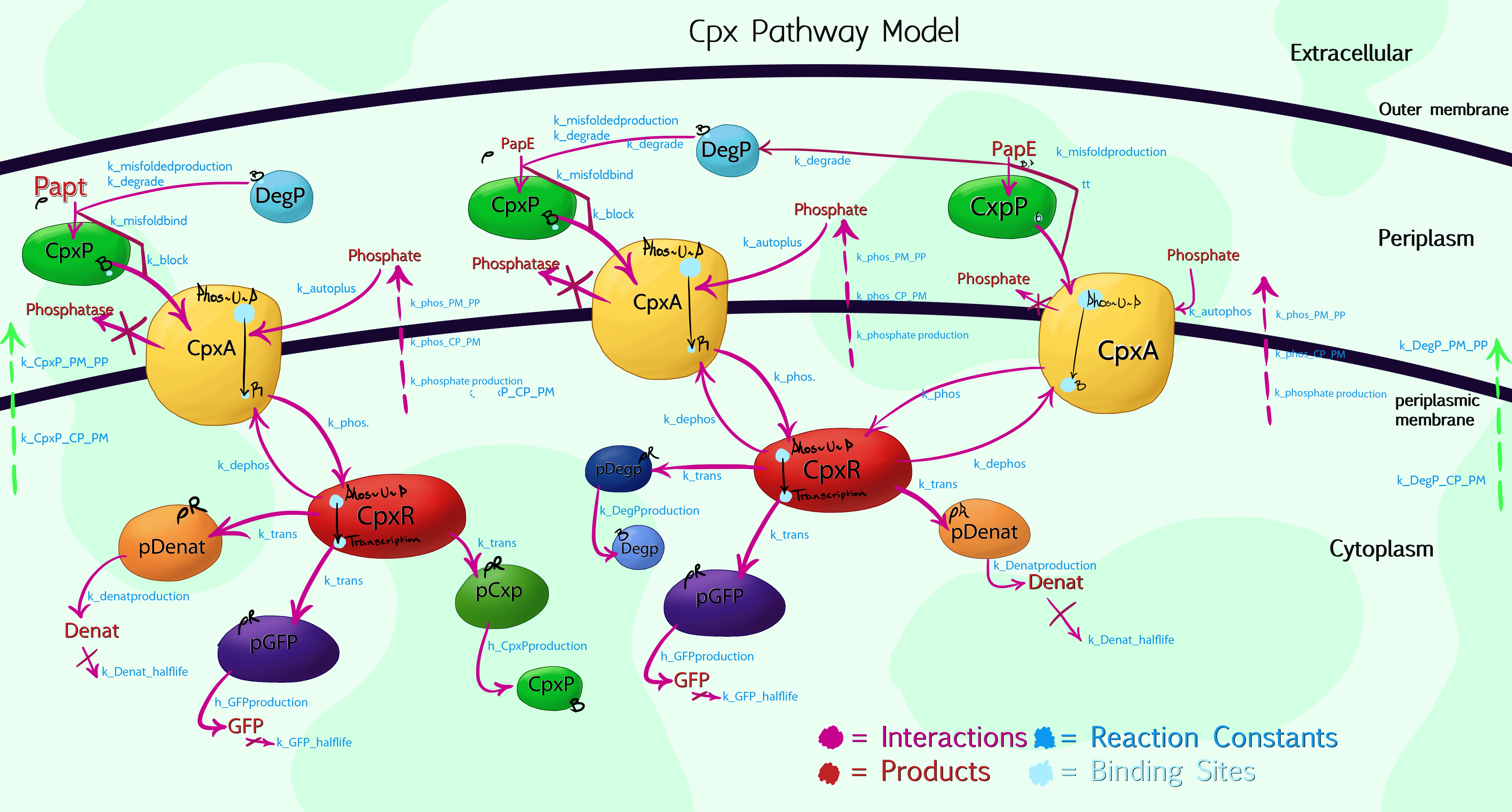
| - | + | [[File:Leeds_modelling.jpeg|centre|800px|A visual model of the Cpx Pathway, click to enlarge|link=https://static.igem.org/mediawiki/2013/5/55/Leeds_modelling.jpeg|frameless]] | |
| + | Due to the high complexity of the Cpx Pathway, it was necessary to graphically plot the processes taking place within the cell by hand, allowing us to then simplify and compartmentalize aspects which, while having an effect on the overall results, were of no interest to us. The above image shows the 3<sup>rd</sup> iteration of this, with colour-coded interactions, products and proteins. The main enzymes of interest were specifically named, while a general product labelled as 'denaturants' was listed to encompass the more than 100 other Cpx-mediated protein syntheses. This would be inaccurate, if not for the use of Rules-Based software, such as RuleBender - for the model to work, one must list the starting concentrations of each molecular species, hence by setting the 'denaturant' promoter concentration to be at least 100 times greater than that of pGFP or pCpxP, the relative interaction mechanics are preserved.<br> | ||
| + | So far, the model is still only at version 0.3, due to the difficulty in confirming the vast number of reaction constants needed - unlike Physics or Chemistry, biological factors will frequently differ by orders of magnitude, dependent upon the conditions they were measured in etc. This is the main downside to using a rule-based approach, as our good friends [[Team:Manchester/Collaboration|Manchester]] and [[Team:York_UK|York]] point out in their modelling support videos. However, the model currently describes the behaviour of CpxP, CpxA, CpxR and the phosphorylation processes these use, as well as the effect of misfolded protein sub-units (caused by membrane stress) and the expected relative production of GFP versus other species produced via CpxR promotion. This should provide a workable, skeletal basis for characterisation of our first [[Team:Leeds/Project#Device I|Bio-System]] as GFP concentration scales directly with fluorescence. As we take measurements, we will then feed this back into the model to fine tune it, and thus develop it further for characterisation of our second [[Team:Leeds/Project#Device 2|system]]. Beyond this, it is hoped the model source code can then be submitted as a beta-stream to the modelling database. It is otherwise available as a [[Media:Leeds_CpxModel_alpha03.txt|raw text document here]] for future teams to make use of and improve upon. | ||
| + | ====Advice on using the model==== | ||
| + | Firstly, it is important to bear in mind that the model is an alpha-version, and not fully optimised or debugged. The code is also written expressly in mind of integration with [http://www.ibiblio.org/virtualcell/ VCell] for visualisation purposes, and makes use of the VCell format for compartmentalisation rules - if you do not wish to view the results within VCell, the syntax will need a large re-write.<br> | ||
| + | Secondly, the code is still awaiting reaction constants, due to large discrepancies in the literature. You are heartily encouraged to improve upon this - regrettably, the team needed to focus on other areas of the [[Team:Leeds/Project|project]] more immeadiately. However, there will hopefully be updates in the future. | ||
| + | ====Assumptions / Justifications==== | ||
| + | The model gets around the problem of not knowing exactly how every reaction progresses in the patway by making specific assumptions: | ||
| + | # The main assumption is that all products of CpxR-induced transcription that are not of interest can be packaged into a singular promoter and product, labelled "denats". The concentration of the promoter (pdenats) must then be set appropriately relative to the remaining promoters - in our case, this should be aound 100 times the amount of pGFP. This allows the reaction mechanics to follow a simplified path, without invalidating the stochastic interaction rates. | ||
| + | # The next most important assumption is that CpxP can only be inhibited by direct binding of a misfolded protein unit. Consequently, the model does not account for the effects of pH, temperature or cellular breakdown upon the components of the pathway. The justification for this is that the Cpx patheway is normally invovled in regulating '''''against''''' these external stress factors, hence we can assume that the metabolites involved are reasonably stable under small fluctuations in these conditions. | ||
| + | # Finally, the model assumes that products such as GFP, denats and phosphates either degrade at constant rates or are consumed in explicitly stated interactions (e.g. phosphorylation). This means we assume that the prescence of these molecules does not affect the rest of the pathway through other regulation mechanisms. This is fair for GFP, as it has an explicit half-life and degradation rate, but is tenuous for the bundled denaturants (they could potentially affect other species involved in the reaction mechanics). However, without thorough agreement in the [http://www.ncbi.nlm.nih.gov/pubmed/11830644 literature] and more direct data measurements, it is impractical to attempt to model these at this stage.<br> | ||
}} | }} | ||
Latest revision as of 03:53, 5 October 2013
| We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project.
Modelling The Cpx PathwayThe key to making MicroBeagle successful hinges on proper integration with the Cpx pathway. As such, it is essential we develop a god working model, not only to predict how much fluorescence we can expect in different environments, but also to prototype and test potential methods for controlling this; in turn reducing our false positive rate.
Cpx Pathway ModelDue to the high complexity of the Cpx Pathway, it was necessary to graphically plot the processes taking place within the cell by hand, allowing us to then simplify and compartmentalize aspects which, while having an effect on the overall results, were of no interest to us. The above image shows the 3rd iteration of this, with colour-coded interactions, products and proteins. The main enzymes of interest were specifically named, while a general product labelled as 'denaturants' was listed to encompass the more than 100 other Cpx-mediated protein syntheses. This would be inaccurate, if not for the use of Rules-Based software, such as RuleBender - for the model to work, one must list the starting concentrations of each molecular species, hence by setting the 'denaturant' promoter concentration to be at least 100 times greater than that of pGFP or pCpxP, the relative interaction mechanics are preserved. Advice on using the modelFirstly, it is important to bear in mind that the model is an alpha-version, and not fully optimised or debugged. The code is also written expressly in mind of integration with [http://www.ibiblio.org/virtualcell/ VCell] for visualisation purposes, and makes use of the VCell format for compartmentalisation rules - if you do not wish to view the results within VCell, the syntax will need a large re-write. Assumptions / JustificationsThe model gets around the problem of not knowing exactly how every reaction progresses in the patway by making specific assumptions:
| |||||||
 |
| ||||||

| |||||||

| |||||||
 "
"