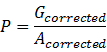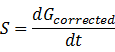Team:ZJU-China/Project/Standardization/Results
From 2013.igem.org
m (→Characterization of Theophylline Riboswitch (BBa_K411003)) |
(→Characterization of Atrazine Riboswitch (BBa_K1054008)) |
||
| (14 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
=== Characterization of Theophylline Riboswitch (BBa_K411003) === | === Characterization of Theophylline Riboswitch (BBa_K411003) === | ||
We choose BBa_K411003, a typical riboswitch used by many iGEM teams, to be our first riboswitch to be standardized, which was firstly designed by iGEM10_NYMU-Taipei and has been tested by ZJU_China 2012. | We choose BBa_K411003, a typical riboswitch used by many iGEM teams, to be our first riboswitch to be standardized, which was firstly designed by iGEM10_NYMU-Taipei and has been tested by ZJU_China 2012. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<center> | <center> | ||
| Line 42: | Line 34: | ||
</center> | </center> | ||
It can be easily observed that the riboswitch’s response to Theophylline is much stronger than to the analogs of theophylline ,including diprophylline, cocaine, caffeine, hypoxanthine and xanthine. | It can be easily observed that the riboswitch’s response to Theophylline is much stronger than to the analogs of theophylline ,including diprophylline, cocaine, caffeine, hypoxanthine and xanthine. | ||
| + | |||
| + | <center> | ||
| + | [[File:K411003_high.png|500px]] | ||
| + | |||
| + | Fig 1.4 Performance Reliability of K411003 (Cultivated with Theophylline) | ||
| + | </center> | ||
| + | |||
| + | <center> | ||
| + | [[File:K411003_low2.png|500px]] | ||
| + | |||
| + | Fig 1.5 Performance Reliability of K411003 (Cultivated with NO Theophylline) | ||
| + | </center> | ||
| + | |||
| + | It obvious that the K411003 riboswitch cannot response correctly to theophylline after being cultivated and diluted for 4 days. | ||
=== Characterization of Atrazine Riboswitch (BBa_K1054008) === | === Characterization of Atrazine Riboswitch (BBa_K1054008) === | ||
| - | + | Our second choice to be standardized is Atrazine Riboswitch. | |
| - | + | <center> | |
| - | + | [[File:Static_of_Atrazine.png|500px]] | |
| - | + | ||
| - | + | Fig 2.1 Static Performance of Atrazine Riboswitch | |
| - | + | </center> | |
| - | + | ||
<center> | <center> | ||
[[File:ZJU-Standardization 2.1.jpg|500px]] | [[File:ZJU-Standardization 2.1.jpg|500px]] | ||
| - | Fig 2. | + | Fig 2.2 Graphic Model about how RFU/OD changes at different time and different concentration of Atrzine. |
</center> | </center> | ||
<center> | <center> | ||
| - | [[File: | + | [[File:Input_of_atrazine.jpg|500px]] |
| - | Fig 2. | + | Fig 2.3 Input Compatibility of Atrazine. |
</center> | </center> | ||
| Line 68: | Line 73: | ||
Various mechanisms about how the aptamer response to inducer have been discovered and we find out a special riboswitch ,which acts differently from K411003. | Various mechanisms about how the aptamer response to inducer have been discovered and we find out a special riboswitch ,which acts differently from K411003. | ||
| - | + | <center> | |
| - | + | [[File:Static_of_Theo(Helix_Spin).png|500px]] | |
| - | + | ||
| - | + | Fig 3.1 Static Performance of Theophylline Riboswitch 2. | |
| - | + | </center> | |
| - | + | ||
| + | <center> | ||
| + | [[File:Dynamic_of_Theo_Helix_Spin.png|500px]] | ||
| + | |||
| + | Fig 3.2 Graphic Model about how GFP/OD changes at different time and different concentration of Theophylline. | ||
| + | </center> | ||
<center> | <center> | ||
| - | [[File: | + | [[File:Theo_high2.png|500px]] |
| - | Fig 3. | + | Fig 3.3 Performance Reliability of Theophylline Riboswitch 2 (Cultivated with Theophylline). |
</center> | </center> | ||
<center> | <center> | ||
| - | [[File: | + | [[File:Theo_low.png|500px]] |
| - | Fig 3. | + | Fig 3.4 Performance Reliability of Theophylline Riboswitch 2 (Cultivated with NO Theophylline). |
</center> | </center> | ||
| Line 107: | Line 117: | ||
'''(4).Plot''' | '''(4).Plot''' | ||
| - | + | === Protocol === | |
| + | '''Protocol for Static & Dynamic Performance''' | ||
| + | # Cultivate the BL21 containing riboswitch in LB at 37℃180rpm until OD600 reaches 0.5. | ||
| + | # Induce the bacteria with 1mM IPTG (final concentration), and cultivate them at 37℃180rpm for 4hrs. | ||
| + | # Dilute the suspension culture with LB to 0.1OD600 . Inducer is added at concentration ranging from 1.00E-05 mol/L(for K411003) or 1.00E-06 mol/L(for Atrazine and Theophylline riboswitch 2)to 1.00E-01 mol/L. | ||
| + | # Measure the RFU ( ex488/em525) and OD600 synchronously. | ||
| + | # Data Analysis | ||
| + | |||
| + | '''Protocol for Performance Reliability''' | ||
| + | # Cultivate the BL21 containing riboswitch in LB at 37℃180rpm for 15hrs. | ||
| + | # The culture was diluted 1:500 into two identical 5mL cultures. Add inducers to one of the cultures to an input level of 1)5mM(for K511003) 2)1mM(for Atrazine and Theophylline Riboswitch 2). | ||
| + | # The culture was diluted 1:500 in the morning and evening every day for 4 days. | ||
| + | # Each evening, a second culture was inoculated by a 1:500 dilution from the culture for both the high and low input condition. These cultures were grown in the absence of inducers for 8hrs to eliminate the accumulated GFP before assaying performance. | ||
| + | # Samples from both of the second copies were induced with inducers at 37℃180rpm for 45 min. Single-cell fluorescence measurements were carried out on a flow cytometer. | ||
| - | Limited by | + | Limited by time and resources, it is impossible for us to characterize all riboswitches. Any teams or individuals are warmly welcomed to enrich our riboswitch standardization library. |
Latest revision as of 19:47, 28 October 2013
Characterization: Results
Contents |
Characterization of Theophylline Riboswitch (BBa_K411003)
We choose BBa_K411003, a typical riboswitch used by many iGEM teams, to be our first riboswitch to be standardized, which was firstly designed by iGEM10_NYMU-Taipei and has been tested by ZJU_China 2012.
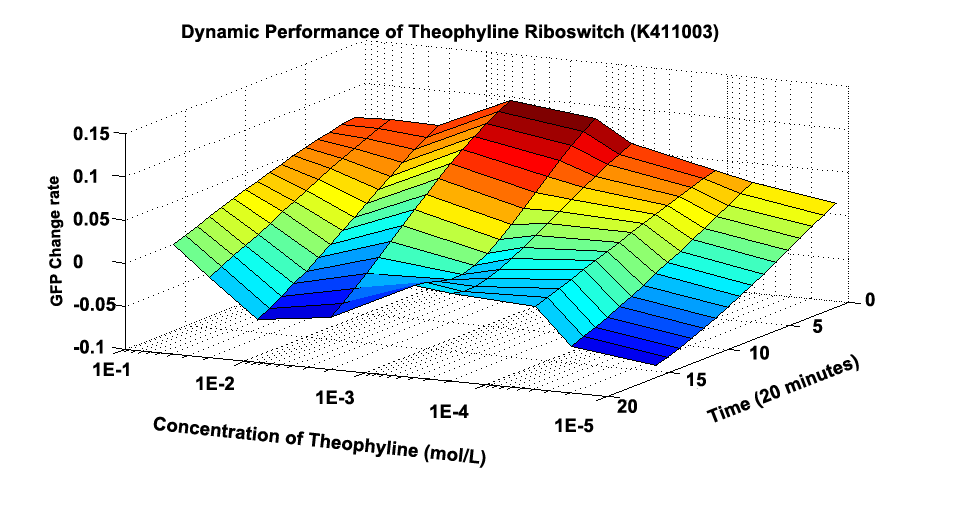
Fig 1.1 shows how the whole GFP Change Rate(GCR) changes at different time and under different concentration. Initially, the culture induced by 0.5M Theophylline has the largest whole GCR. And almost every culture’s GCR experiences decrement along time.
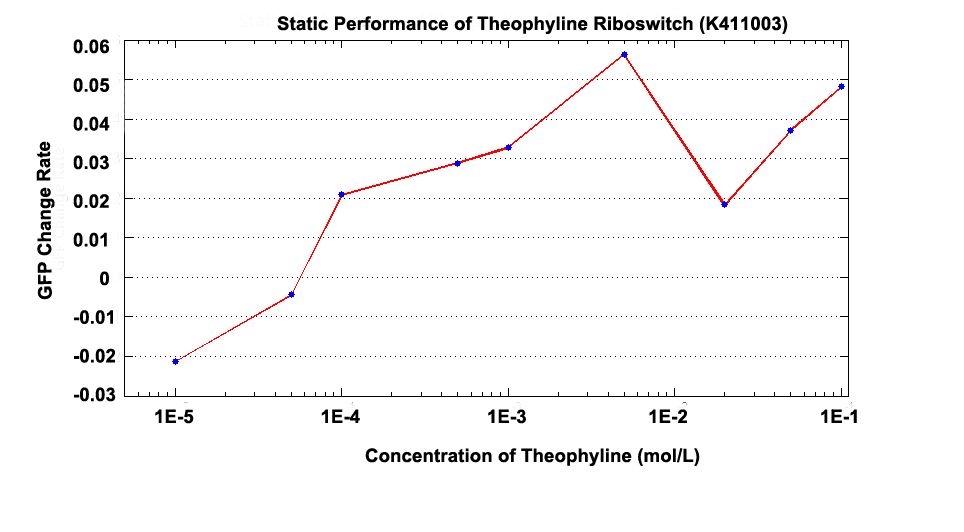
Fig 1.2 GFP Change Rate versus Concentration at 100 minutes after Theophylline was added.
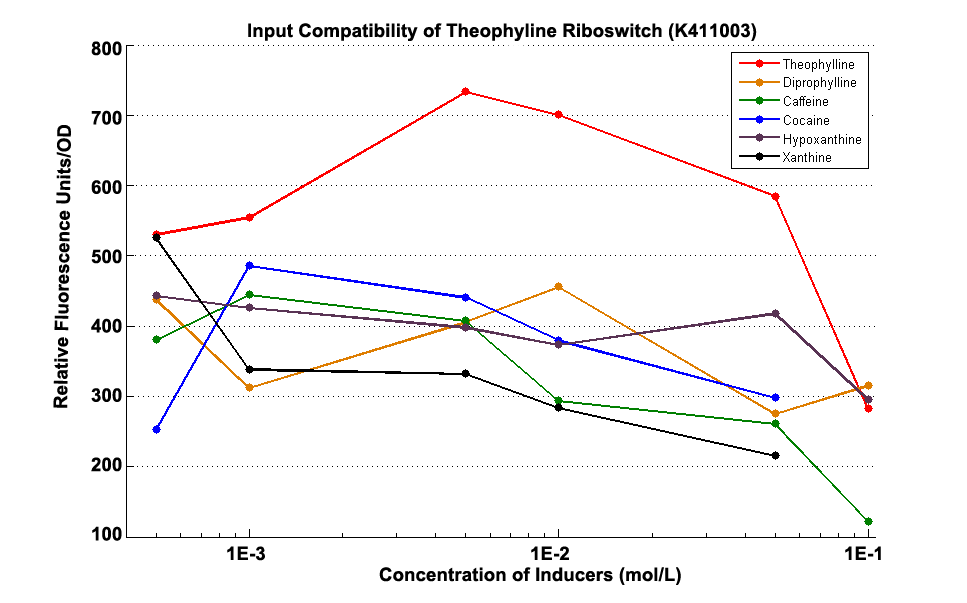
Fig 1.3 Input Compatibility of K411003
It can be easily observed that the riboswitch’s response to Theophylline is much stronger than to the analogs of theophylline ,including diprophylline, cocaine, caffeine, hypoxanthine and xanthine.
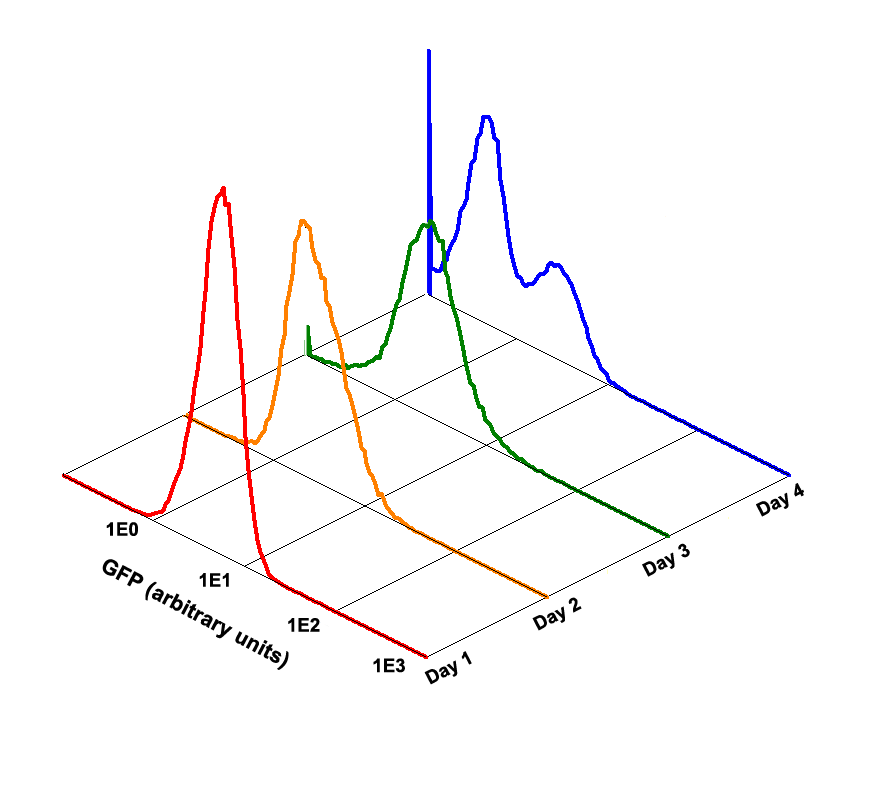
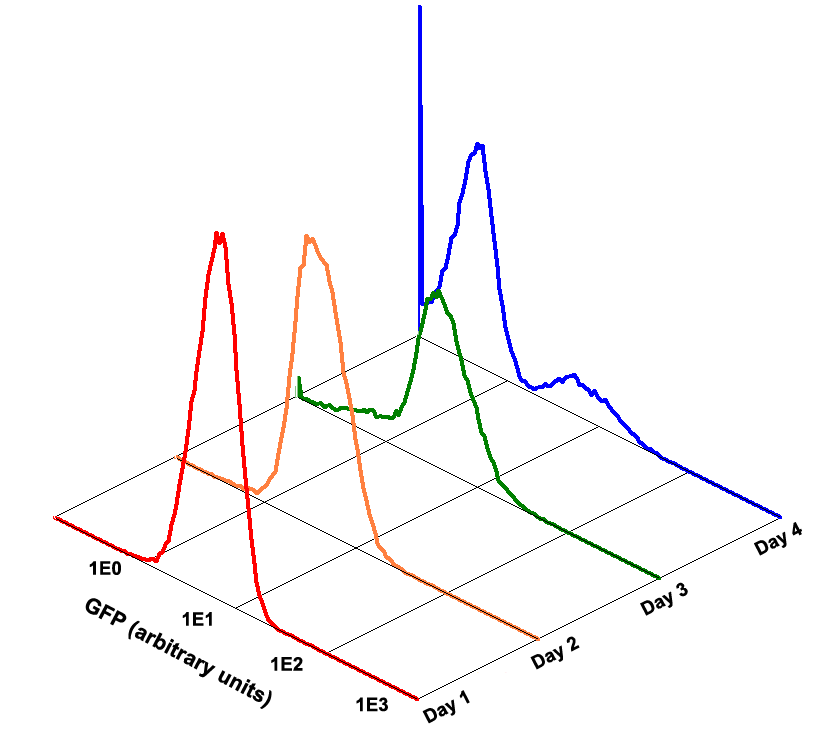
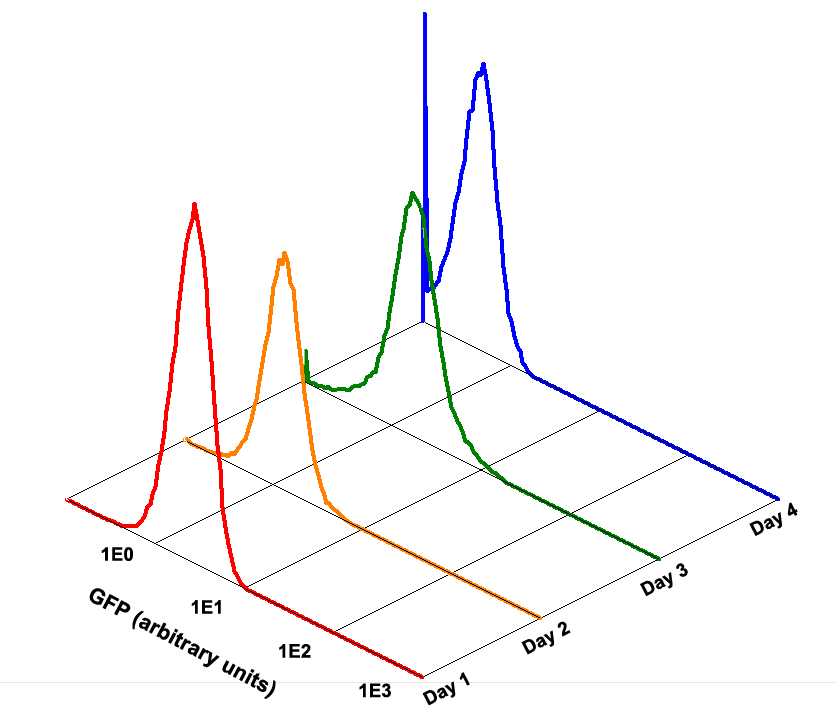
Fig 1.4 Performance Reliability of K411003 (Cultivated with Theophylline)
Fig 1.5 Performance Reliability of K411003 (Cultivated with NO Theophylline)
It obvious that the K411003 riboswitch cannot response correctly to theophylline after being cultivated and diluted for 4 days.
Characterization of Atrazine Riboswitch (BBa_K1054008)
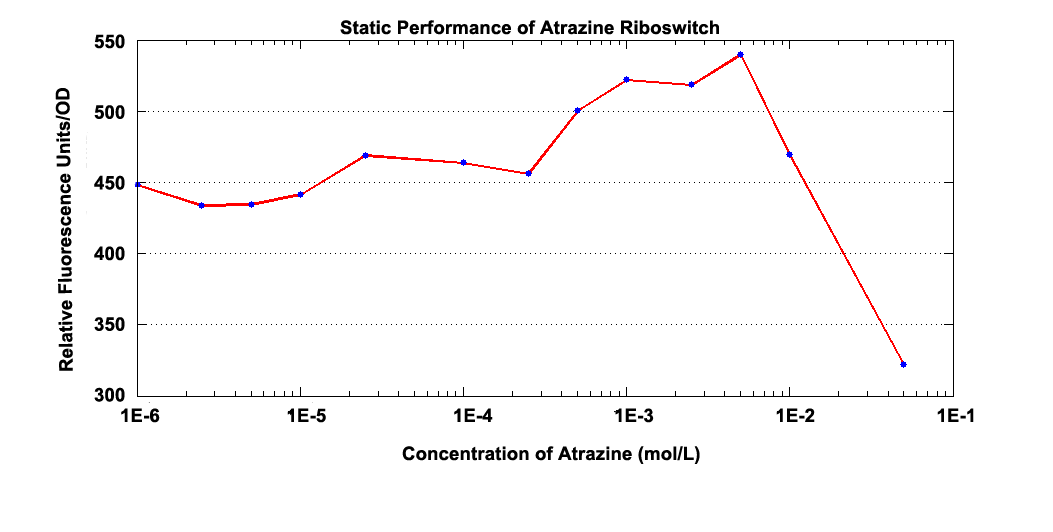
Our second choice to be standardized is Atrazine Riboswitch.
Fig 2.1 Static Performance of Atrazine Riboswitch
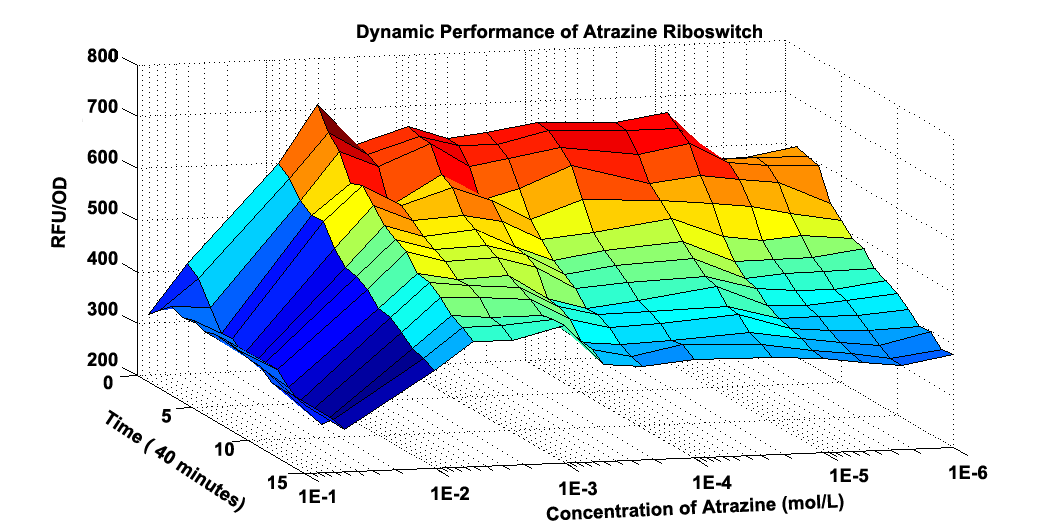
Fig 2.2 Graphic Model about how RFU/OD changes at different time and different concentration of Atrzine.
Fig 2.3 Input Compatibility of Atrazine.
Characterization of Theophylline Riboswitch 2 (BBa_K1054022 )
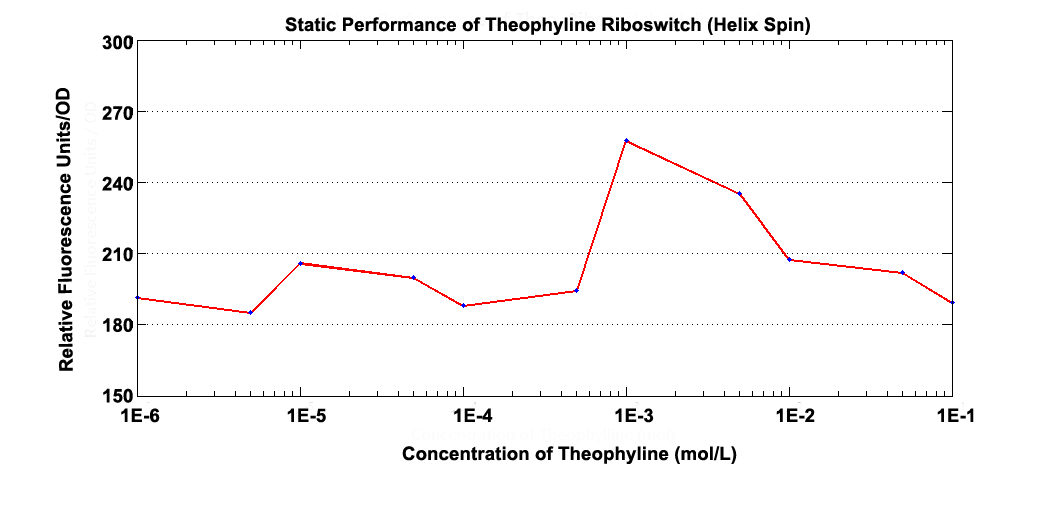
Various mechanisms about how the aptamer response to inducer have been discovered and we find out a special riboswitch ,which acts differently from K411003.
Fig 3.1 Static Performance of Theophylline Riboswitch 2.
Fig 3.2 Graphic Model about how GFP/OD changes at different time and different concentration of Theophylline.
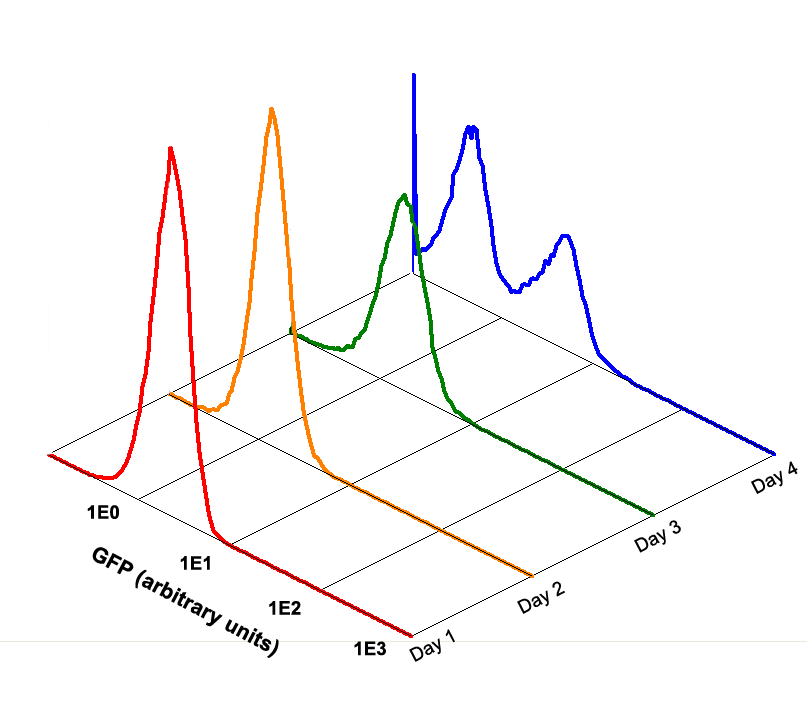
Fig 3.3 Performance Reliability of Theophylline Riboswitch 2 (Cultivated with Theophylline).
Fig 3.4 Performance Reliability of Theophylline Riboswitch 2 (Cultivated with NO Theophylline).
Data Analysis
Theoretically, data processing should consist of two stages including Processing and Calibration, respectively. Lacking enough time and equipment, we cannot finish the Calibration part-which is about to find out the relation about OD600 versus the number of cells and RFU versus mass of GFP-before wiki freeze . Here comes the Processing Part:
(1).Subtract the backgrounds from the raw data
(2).Calculate the RFU of per unit, P
(3).Calculate the whole RFU change rate, S
(4).Plot
Protocol
Protocol for Static & Dynamic Performance
- Cultivate the BL21 containing riboswitch in LB at 37℃180rpm until OD600 reaches 0.5.
- Induce the bacteria with 1mM IPTG (final concentration), and cultivate them at 37℃180rpm for 4hrs.
- Dilute the suspension culture with LB to 0.1OD600 . Inducer is added at concentration ranging from 1.00E-05 mol/L(for K411003) or 1.00E-06 mol/L(for Atrazine and Theophylline riboswitch 2)to 1.00E-01 mol/L.
- Measure the RFU ( ex488/em525) and OD600 synchronously.
- Data Analysis
Protocol for Performance Reliability
- Cultivate the BL21 containing riboswitch in LB at 37℃180rpm for 15hrs.
- The culture was diluted 1:500 into two identical 5mL cultures. Add inducers to one of the cultures to an input level of 1)5mM(for K511003) 2)1mM(for Atrazine and Theophylline Riboswitch 2).
- The culture was diluted 1:500 in the morning and evening every day for 4 days.
- Each evening, a second culture was inoculated by a 1:500 dilution from the culture for both the high and low input condition. These cultures were grown in the absence of inducers for 8hrs to eliminate the accumulated GFP before assaying performance.
- Samples from both of the second copies were induced with inducers at 37℃180rpm for 45 min. Single-cell fluorescence measurements were carried out on a flow cytometer.
Limited by time and resources, it is impossible for us to characterize all riboswitches. Any teams or individuals are warmly welcomed to enrich our riboswitch standardization library.
 "
"