Team:TU-Munich/Results/KillSwitch
From 2013.igem.org
(Difference between revisions)
(→Kill Switch) |
(→Kill Switch) |
||
| Line 29: | Line 29: | ||
* konstrukt zu groß für trafo [homologe regionen??] | * konstrukt zu groß für trafo [homologe regionen??] | ||
* filter nicht optimal [bild filter, ingmar @ blaulicht bench?] | * filter nicht optimal [bild filter, ingmar @ blaulicht bench?] | ||
| + | |||
| + | ==Results== | ||
| + | |||
| + | |||
<div class="visualClear"></div> | <div class="visualClear"></div> | ||
<!-- End of content --> | <!-- End of content --> | ||
Revision as of 21:39, 28 October 2013
Kill Switch
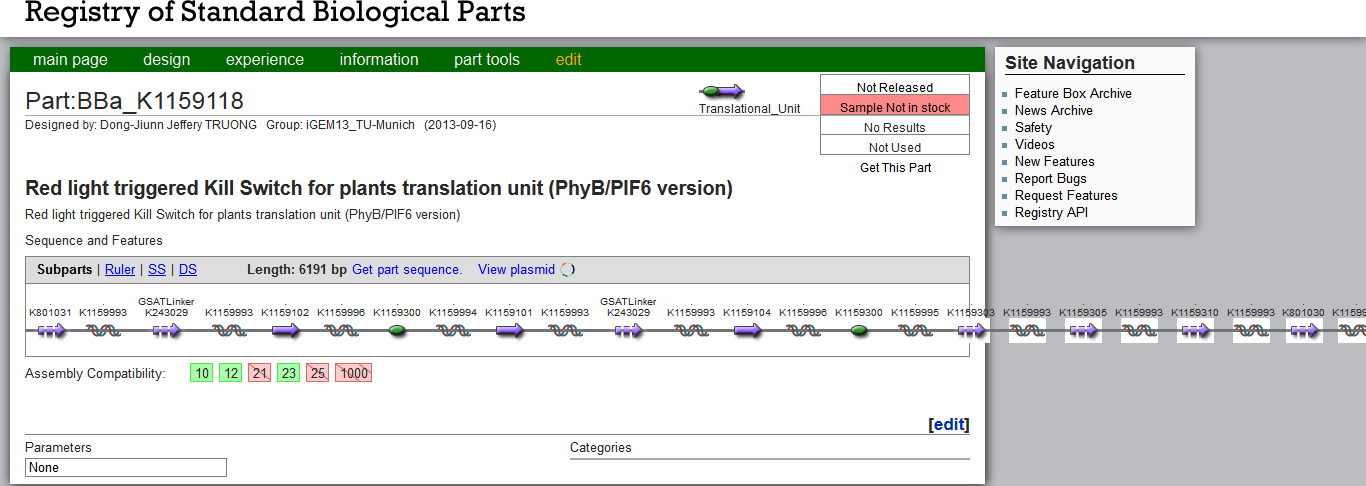
The Kill Switch mechanism is the most complex and ambitious aspect of our project on the protein level. The DNA constructs are very large and even exceed the Part Registry´s frame (Fig. 1). For details on its design and function see here. We were very excited to start
File:PBI 376 f1.gif
Figure 2: Comparative expression performance of different constitutive mammalian and plant promoters in Physcomitrella patens, http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009
[http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2003] "Targeted gene-knockout in Physco" paper
- in bakkis nicht exprimiert [codon usage]
- promotor zu stark [bild vgl prom stärken]
- konstrukt zu groß für trafo [homologe regionen??]
- filter nicht optimal [bild filter, ingmar @ blaulicht bench?]
Results
 "
"




AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351