Team:TU-Delft
From 2013.igem.org
(Difference between revisions)
| (302 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:TU-Delft/Templates/Navigation}} | {{:Team:TU-Delft/Templates/Navigation}} | ||
{{:Team:TU-Delft/Templates/Style}} | {{:Team:TU-Delft/Templates/Style}} | ||
| - | {{:Team:TU-Delft/Templates/ | + | {{:Team:TU-Delft/Templates/Frog}} |
| + | {{:Team:TU-Delft/Templates/Logo}} | ||
<html> | <html> | ||
| - | <div id=" | + | |
| + | <div style="margin-top:-20px;margin-bottom:5px;"> | ||
| + | <center> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/8/82/Pep9.jpg"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/8/82/Pep9.jpg" alt="" HEIGHT=90" WIDTH="700" /> | ||
| + | </a> | ||
| + | </center> | ||
| + | </div> | ||
| + | |||
| + | <!--copyright METU TEAM iGem 2012--> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-type" content="text/html; charset=utf-8" /> | ||
| + | |||
| + | |||
| + | <link rel="stylesheet" href="http://simsekburak.com/hasan/css/style.css" type="text/css" media="all" /> | ||
| + | <!--[if IE 6]> | ||
| + | <link rel="stylesheet" href="css/ie6.css" type="text/css" media="all" /> | ||
| + | <script src="http://simsekburak.com/hasan/js/png-fix.js" type="text/javascript"></script> | ||
| + | <![endif]--> | ||
| + | <script src="http://simsekburak.com/hasan/js/jquery-1.4.2.js" type="text/javascript"></script> | ||
| + | <script src="http://simsekburak.com/hasan/js/jquery.jcarousel.js" type="text/javascript"></script> | ||
| + | <script src="http://simsekburak.com/hasan/js/js-func.js" type="text/javascript"></script> | ||
| + | |||
| + | <style> | ||
| + | body{ | ||
| + | background:url("https://static.igem.org/mediawiki/2013/3/32/BackroundDelft.jpg"); | ||
| + | background-repeat:no-repeat; | ||
| + | border-top: none; | ||
| + | |||
| + | } | ||
| + | |||
| + | .slider-holder{ | ||
| + | |||
| + | width:964px; | ||
| + | } | ||
| + | .slider-left ul li h2 { | ||
| + | |||
| + | margin-left:10px; | ||
| + | } | ||
| + | .slider-right{ | ||
| + | width:610px; | ||
| + | } | ||
| + | |||
| + | h8{ | ||
| + | margin-left:10px; | ||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | #header{ | ||
| + | height: 150px; | ||
| + | } | ||
| + | </style> | ||
| + | |||
| + | </head> | ||
| + | <!-- Slider --> | ||
| + | <div id="slider"> | ||
| + | <div class="shell"> | ||
| + | <div class="slider-holder"> | ||
| + | <div class="slider-left"> | ||
| + | <ul> | ||
| + | <li> | ||
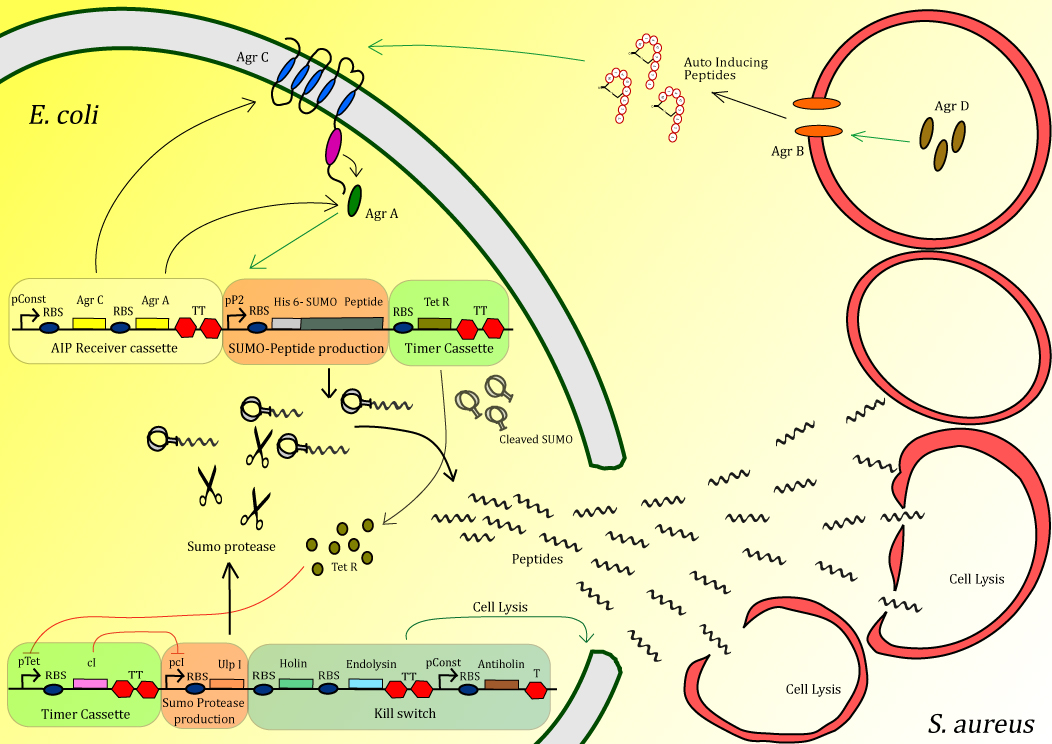
| + | <h2>Who are we?</h2><div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify">We are the iGEM 2013 team of Delft University of Technology, Netherlands. We are working on the production of antimicrobial peptides in order to kill methicillin resistant <i>Staphylococcus aureus, MRSA</i>.</p> | ||
| + | |||
| + | </li> | ||
| + | |||
| + | <li> <h2>Aim</h2><div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify"> To engineer <i>Escherichia coli</i> that can detect MRSA in order to locally produce and deliver antimicrobial peptides. The project is divided into several essential modules. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <h2>Sensing autoinducing peptides</h2> | ||
| + | <div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify">The sensing part of the of <i>Staphylococcus aureus</i> Agr system is engineered in <i>E. coli</i>. In that way <i>E. coli</i> detects the AIPs that are produced by <i>Staphylococcus aureus</i> which induces the peptide production. </p> | ||
| + | |||
| + | |||
| + | </li> | ||
| + | <li> | ||
| + | <h2>Peptide Production</h2> | ||
| + | <div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify"> <i>E. coli</i> detects <i>Staphylococcus aureus</i> by its own quorum sensing system, and at that moment inactivated antimicrobial peptides are being produced </p> | ||
| + | </li> | ||
| + | <li> | ||
| + | <h2>Timer</h2> | ||
| + | <div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify"> | ||
| + | With the use of a timer the peptide will be activated by cleavage of an inactivating tag. With the use of a timer, high concentrations of peptide can be delivered very locally in order to efficiently kill MRSA.</p> | ||
| + | |||
| + | </li> | ||
| + | <li> | ||
| + | <h2>Kill Switch</h2> | ||
| + | <div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify"> | ||
| + | In order to ensure the safety of the system, the peptide activating protease is combined with a kill switch that will kill our E. coli.</p> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <h2>Zephyr</h2> | ||
| + | <div style="margin-left:10px;margin-right:10px;float:left;display:inline-block;"><p align="justify"> | ||
| + | Zephyr is a low-cost Do It Yourself (DIY) machine which can scan petridishes and 96 well plates for expression of fluorescence at micrometer scale. | ||
| + | </p> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="slider-right"> | ||
| + | <ul> | ||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Team"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/7/7f/Team_Delft1.jpg" alt="" HEIGHT="348" WIDTH="600" /> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/ProjectOverview"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/5/51/Schem-new.jpg" alt="" HEIGHT="348" WIDTH="600"/> | ||
| + | </a> | ||
| + | |||
| + | </li> | ||
| + | |||
| + | |||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Sensing"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/6/6d/Sensing-AIP.jpg" alt="" HEIGHT="348" WIDTH="600"/> | ||
| + | </a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Peptides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/f/fe/Peptide-2.jpg" alt="" HEIGHT="348" WIDTH="600"/> | ||
| + | </a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Timer"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/3/34/Timer-2.jpg" alt="" HEIGHT="348" WIDTH="600"/> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Killswitch"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/f/f8/Kill2.jpg" alt="" HEIGHT="348" WIDTH="600"/> | ||
| + | </a></li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Zephyr"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/4/4c/2D_table_ZephyrHome.jpg" alt="" WIDTH="600"/> | ||
| + | </a> | ||
| + | |||
| + | </li> | ||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="cl"> </div> | ||
| + | <div class="slider-nav"> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Team">1</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/ProjectOverview">2</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Sensing">3</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Peptides">4</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Timer">5</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Killswitch">6</a> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Zephyr">7</a> | ||
| + | |||
| + | <div class="cl"> </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!-- End Slider --> | ||
| + | <!-- Main --> | ||
| + | <br> | ||
| + | |||
| + | <div style="margin-left:40px;width:420px;float:left;display:inline-block;"> | ||
| + | <br> | ||
| + | |||
| + | |||
<script>(function(d, s, id) { | <script>(function(d, s, id) { | ||
var js, fjs = d.getElementsByTagName(s)[0]; | var js, fjs = d.getElementsByTagName(s)[0]; | ||
| Line 12: | Line 187: | ||
js.src = "//connect.facebook.net/en_US/all.js#xfbml=1"; | js.src = "//connect.facebook.net/en_US/all.js#xfbml=1"; | ||
fjs.parentNode.insertBefore(js, fjs); | fjs.parentNode.insertBefore(js, fjs); | ||
| - | }(document, 'script', 'facebook-jssdk'));</script> | + | }(document, 'script', 'facebook-jssdk'));</script> |
| - | + | <div class="fb-like-box" data-href="https://www.facebook.com/pages/TUDelft-iGEM-2013/627491607266909?fref=ts" data-width="380" data-height="300" data-show-faces="false" data-stream="true" data-show-border="true" data-header="true"> | |
| - | <div class="fb-like-box" data-href="https://www.facebook.com/pages/TUDelft-iGEM-2013/627491607266909?fref=ts" data-width=" | + | |
</div> | </div> | ||
| + | <br> | ||
| + | <a href="http://www2.clustrmaps.com/user/9f010c206"><img src="http://www2.clustrmaps.com/stats/maps-no_clusters/2013.igem.org-Team-TU-Delft-thumb.jpg" alt="Locations of visitors to this page" /> | ||
| + | </a> | ||
| + | <a href="http://www.easycounter.com/"> | ||
| + | <img src="http://www.easycounter.com/counter.php?tudelft3" | ||
| + | border="0" alt="Free Web Counters"></a> | ||
| + | </div> | ||
| + | |||
| + | <div style="margin-left:50px;width:200px;display:inline-block;"> | ||
| + | <object width="420" height="315"><param name="movie" value="//www.youtube.com/v/jMUHYKS2jvc?hl=el_GR&version=3"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="//www.youtube.com/v/jMUHYKS2jvc?hl=el_GR&version=3" type="application/x-shockwave-flash" width="420" height="315" allowscriptaccess="always" allowfullscreen="true"></embed></object> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
<center> | <center> | ||
| - | < | + | <p> <h3> Check out what news in Netherlands has to say about the achievements of our Team </p><a href="https://2013.igem.org/Team:TU-Delft/PR" target="blank"> In News. </a> </h3> <br> </center> |
| - | </center> | + | |
| - | <div style="margin- | + | <div style="margin-top:10px;margin-left:400px;float:left;display:inline-block;"> |
| + | <h2>Our sponsors</h2> | ||
| + | <hr> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <center> | ||
| + | <table style="border-spacing: 3px; alignment: center;" cellspacing="0" cellpadding="0"> | ||
| + | <tr > | ||
| + | <td colspan="4" style="text-align: center;"><h3></h3></td> | ||
| + | </tr> | ||
| - | < | + | <tr> |
| + | <!--<td style="text-align: center;" width="250px"> | ||
| + | <a href="http://www.bt.tudelft.nl/" target="_blank"><img src="https://static.igem.org/mediawiki/2012/thumb/e/e6/TUD_BT.png/180px-TUD_BT.png" width=177 height=60></a> | ||
| + | |||
| + | </td>--> | ||
| + | |||
| + | <!--BT--> <td style="text-align: center;" width="250px"> | ||
| + | <a href="http://home.tudelft.nl/en" target="_blank"><img src="https://static.igem.org/mediawiki/igem.org/5/5a/Logotudelft.png" width=215 height=60></a> | ||
| - | < | + | </td> |
| - | </ | + | <!--BN--> <td style="text-align: center;" width="250px"> |
| + | |||
| + | <a href="http://www.bt.tudelft.nl/" target="_blank"><img src="https://static.igem.org/mediawiki/2012/thumb/e/e6/TUD_BT.png/180px-TUD_BT.png" width=177 height=60></a> | ||
| + | </td> | ||
| + | </tr> | ||
| - | < | + | <tr> |
| + | <td style="text-align: center;" width="250px"> | ||
| + | <!--dcsc--> | ||
| + | <a href="http://www.tnw.tudelft.nl" target="_blank"><img src="https://static.igem.org/mediawiki/2012/thumb/0/02/TUD_BioNanoScience.jpg/180px-TUD_BioNanoScience.jpg" width=276 height=60></a> | ||
| + | </td> | ||
| + | <td style="text-align: center;" width="250px"> | ||
| + | <!--Bio--> <a href="http://bioinformatics.tudelft.nl/" target="_blank"><img src="https://static.igem.org/mediawiki/2012/thumb/4/4e/TUD_BioInformatics.png/180px-TUD_BioInformatics.png" width=144 height=60> </a> | ||
| + | </td> | ||
| + | </tr> | ||
| - | < | + | <tr> |
| - | + | <td style="text-align: center;" width="250px"> | |
| + | <a href="http://www.be-basic.org/fileadmin/custom/images/logo-new.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/9/9b/Bebasic.png" width=88 height=60> </a> | ||
| + | </td> | ||
| + | <td style="text-align: center;" width="250px"> | ||
| + | <a href="http://stud.nl/" target="_blank"><img src="https://static.igem.org/mediawiki/2012/thumb/7/75/TUD_stuD.jpg/180px-TUD_stuD.jpg" width=136 height=60> </a> | ||
| + | </td> | ||
| + | <td style="text-align: center;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="text-align: center;" width="250px"> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/archive/5/58/20131028223200!Logo_ERASynBio.png"target="_blank"><img src=" https://static.igem.org/mediawiki/2013/archive/5/58/20131028223200!Logo_ERASynBio.png"width=120 height=40> </a> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
| + | </div> | ||
| - | |||
| - | |||
| - | < | + | <!-- End Main --> |
| - | + | ||
</html> | </html> | ||
Latest revision as of 22:37, 28 October 2013

 "
"