Team:ZJU-China/Project/GhostSensor/Background
From 2013.igem.org
m (→E-lysis and bacterial ghost) |
(→Split TEM-1 β-lactamase) |
||
| (12 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<div class="content"> | <div class="content"> | ||
</html> | </html> | ||
| - | == Ghost | + | == Bacterial Ghost: Background == |
__TOC__ | __TOC__ | ||
| - | |||
| - | |||
| - | |||
=== Aptamers === | === Aptamers === | ||
Aptamers are functional oligonucleotides that could specifically bind to target molecules and thus are termed chemical antibodies. After nearly 20 years’ endeavor, DNA and RNA aptamers have been identified as binding tightly to a wide range of targets including proteins, peptides, amino acids, drugs, metal ions and even whole cells. | Aptamers are functional oligonucleotides that could specifically bind to target molecules and thus are termed chemical antibodies. After nearly 20 years’ endeavor, DNA and RNA aptamers have been identified as binding tightly to a wide range of targets including proteins, peptides, amino acids, drugs, metal ions and even whole cells. | ||
| - | Aptamer-based sensors possess unprecedented advantages compared to biosensors based on natural receptors or antibodies for the following reasons | + | Aptamer-based sensors possess unprecedented advantages compared to biosensors based on natural receptors or antibodies for the following reasons: |
| + | # They can in principle be selected for any given target ranging from small molecules to the whole cells. | ||
| + | # They can be synthesized with high reproducibility and purity from commercial sources at a low cost which also make them the ideal molecule for gene engineering. | ||
| + | # DNA aptamers are usually more chemically stable than antibodies or enzymes. | ||
By far, there have been numerous aptamer-based biosensors based on a variety of mechanisms. However, most of them are confined to detect small molecules and strongly rely on the conformational change of aptamers upon binding to targets. ZJU-China this year want to find a quick and robust way to detect large molecules independent of aptamer conformational status. | By far, there have been numerous aptamer-based biosensors based on a variety of mechanisms. However, most of them are confined to detect small molecules and strongly rely on the conformational change of aptamers upon binding to targets. ZJU-China this year want to find a quick and robust way to detect large molecules independent of aptamer conformational status. | ||
| - | The idea is simply two things: membrane scaffolds and dimerization. Two aptamers recognizing two different sites of the same target molecule can pull membrane scaffolds together and lead to dimerization. A split GFP or enzyme fused to membrane scaffolds will then come to a whole again and give signal output. However, when it comes to practical design, we have to tackle a series of problems. | + | The idea is simply two things: membrane scaffolds and dimerization. Two aptamers recognizing two different sites of the same target molecule can pull membrane scaffolds together and lead to dimerization. A split GFP or enzyme fused to membrane scaffolds will then come to a whole again and give signal output. However, when it comes to practical design, we have to tackle a series of problems. |
=== E-lysis and bacterial ghost === | === E-lysis and bacterial ghost === | ||
| Line 33: | Line 33: | ||
<center> | <center> | ||
[[File:Ghostpaperimage.jpg|600px|center]] | [[File:Ghostpaperimage.jpg|600px|center]] | ||
| - | + | </center><center> | |
| - | Electronic microscopy of bacterial ghost. (A. Witte, et al., Dynamics of PhiX174 protein E-mediated lysis of ''Escherichia coli'', Arch Microbiol (1992) 157:381 388) | + | Electronic microscopy of bacterial ghost. <br> |
| - | + | <small>(A. Witte, et al., Dynamics of PhiX174 protein E-mediated lysis of ''Escherichia coli'', Arch Microbiol (1992) 157:381 388)</small> | |
| - | + | ||
</center> | </center> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Biotin-avidin interaction === | === Biotin-avidin interaction === | ||
| Line 77: | Line 63: | ||
</div> | </div> | ||
<div style="float:right;">Next: | <div style="float:right;">Next: | ||
| - | <a href="./Design">Ghost | + | <a href="./Design">Bacterial Ghost: Design</a> |
</div> | </div> | ||
Latest revision as of 01:37, 29 October 2013
Bacterial Ghost: Background
Contents |
Aptamers
Aptamers are functional oligonucleotides that could specifically bind to target molecules and thus are termed chemical antibodies. After nearly 20 years’ endeavor, DNA and RNA aptamers have been identified as binding tightly to a wide range of targets including proteins, peptides, amino acids, drugs, metal ions and even whole cells.
Aptamer-based sensors possess unprecedented advantages compared to biosensors based on natural receptors or antibodies for the following reasons:
- They can in principle be selected for any given target ranging from small molecules to the whole cells.
- They can be synthesized with high reproducibility and purity from commercial sources at a low cost which also make them the ideal molecule for gene engineering.
- DNA aptamers are usually more chemically stable than antibodies or enzymes.
By far, there have been numerous aptamer-based biosensors based on a variety of mechanisms. However, most of them are confined to detect small molecules and strongly rely on the conformational change of aptamers upon binding to targets. ZJU-China this year want to find a quick and robust way to detect large molecules independent of aptamer conformational status.
The idea is simply two things: membrane scaffolds and dimerization. Two aptamers recognizing two different sites of the same target molecule can pull membrane scaffolds together and lead to dimerization. A split GFP or enzyme fused to membrane scaffolds will then come to a whole again and give signal output. However, when it comes to practical design, we have to tackle a series of problems.
E-lysis and bacterial ghost
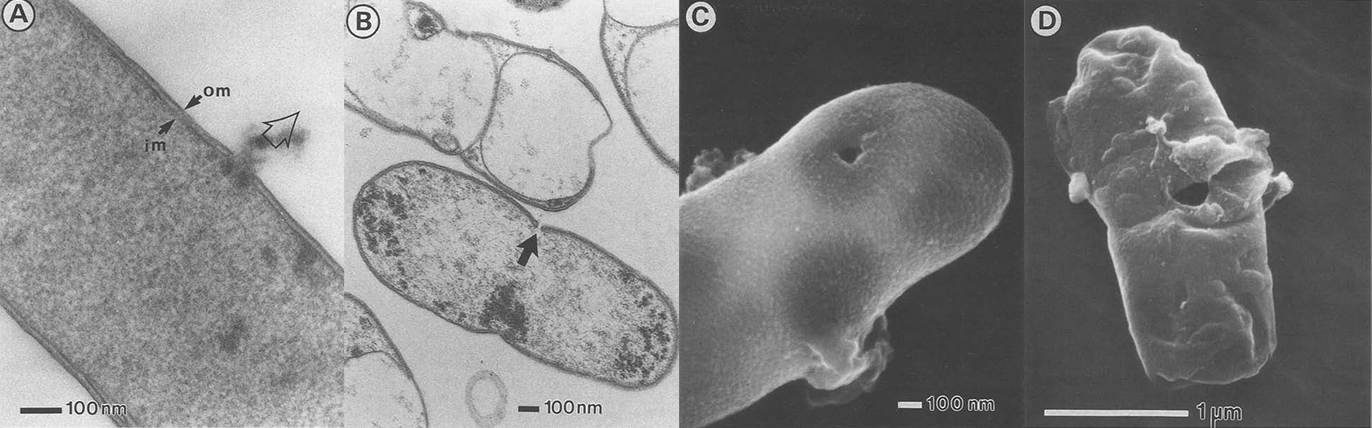
Because we choose E. coli as our chassis, we don’t want to express our membrane scaffolds in the outer-membrane due to its rigidity and scarcity of proteins. As a result, we express our protein scaffolds in the inner-membrane and make tunnels through inner- and outer-membrane to let target protein freely diffuse into the cell. By this way, we have to make sure that certain level of integrity must be retained or elsewise the tunneled-cells will breakdown into pieces. So we choose to use a simple method for opening cells by PhiX174 protein E-mediated lysis (E-lysis). This lysis procedure is very gentle, avoiding all undesireable side effects of mechanical or chemical procedures to destroy the cell envelope. E-lysis introduces a transmembrane tunnel structure with a diameter of 40–200 nm in the envelope complex of the cells. The hole has fused inner and outer membranes at its orifice, and the driving force for E-lysis is the osmotic pressure difference between the cytoplasm and the medium.
There are two great things about E-lysis. First, it keeps the periplasmic space intact which permits further use of this microenvironment. Second, we can adjust the diameter of the holes simply by including different concentrations of MgSO4 into the solution, which allow different size of targets ranging from proteins to virus to come in.
The resultant cell envelope is called bacterial ghost. The bacterial ghost platform technology has been used as an innovative system for vaccine, drug or active substance delivery. Using bacterial ghosts as delivery vehicles for subunit or DNA-vaccines the particle structure and surface properties of bacterial ghosts are targeting the carrier itself to primary antigen-presenting cells. Here we render bacterial ghosts a new role as an ideal candidate for membrane scaffold-based biosensor.
Electronic microscopy of bacterial ghost.
(A. Witte, et al., Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli, Arch Microbiol (1992) 157:381 388)
Biotin-avidin interaction
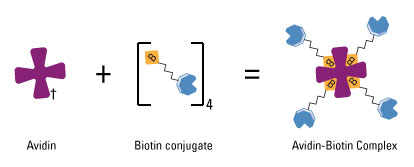
Biotin-avidin interaction has a dissociation constant Kd on the order of 10−15 M, which is one of the strongest known protein-ligand interactions. Biotin is a small chemical which can be modified to DNA or RNA. By biotinylation of our aptamers, we can link aptamers (which is oligonucleotides) to proteins via biotin-avidin interaction. In order to achieve this goal, we fused streptavidin, which is a commonly used tetramer avidin, to the cytoplasmic terminal of our inner-membrane scaffold. So here our aptamer acts like a bridge with its one end attached to target molecules and the other end firmly hold to membrane scaffolds.
Schem illustrating the avidin-biotin interaction. Avidin, streptavidin or NeutrAvidin Protein can bind up to four biotin molecules, which are normally conjugated to an enzyme, antibody or target protein to form an avidin-biotin complex. (Thermo Scientific)
References
[1] Juewen Liu, Zehui Cao, and Yi Lu, Functional Nucleic Acid Sensors, Chem. Rev. 2009, 109, 1948–1998
[2] S. Resch et al., Aqueous release and purification of poly(b-hydroxybutyrate) from Escherichia coli, Journal of Biotechnology 65 (1998) 173–182
[3] Hijiri Hasegawa et al., Improvement of Aptamer Affinity by Dimerization, Sensors 2008, 8, 1090-1098
[4] André Galarneau et al., β-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein–protein interactions, nature biotechnology 2002, 20, 619-22
 "
"