Team:ZJU-China/Project/GhostSensor/Design
From 2013.igem.org
(→Ghost Sensor: Design) |
(→Bacterial Ghost: Design) |
||
| (25 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<div class="content"> | <div class="content"> | ||
</html> | </html> | ||
| - | == Ghost | + | == '''Bacterial Ghost: Design''' == |
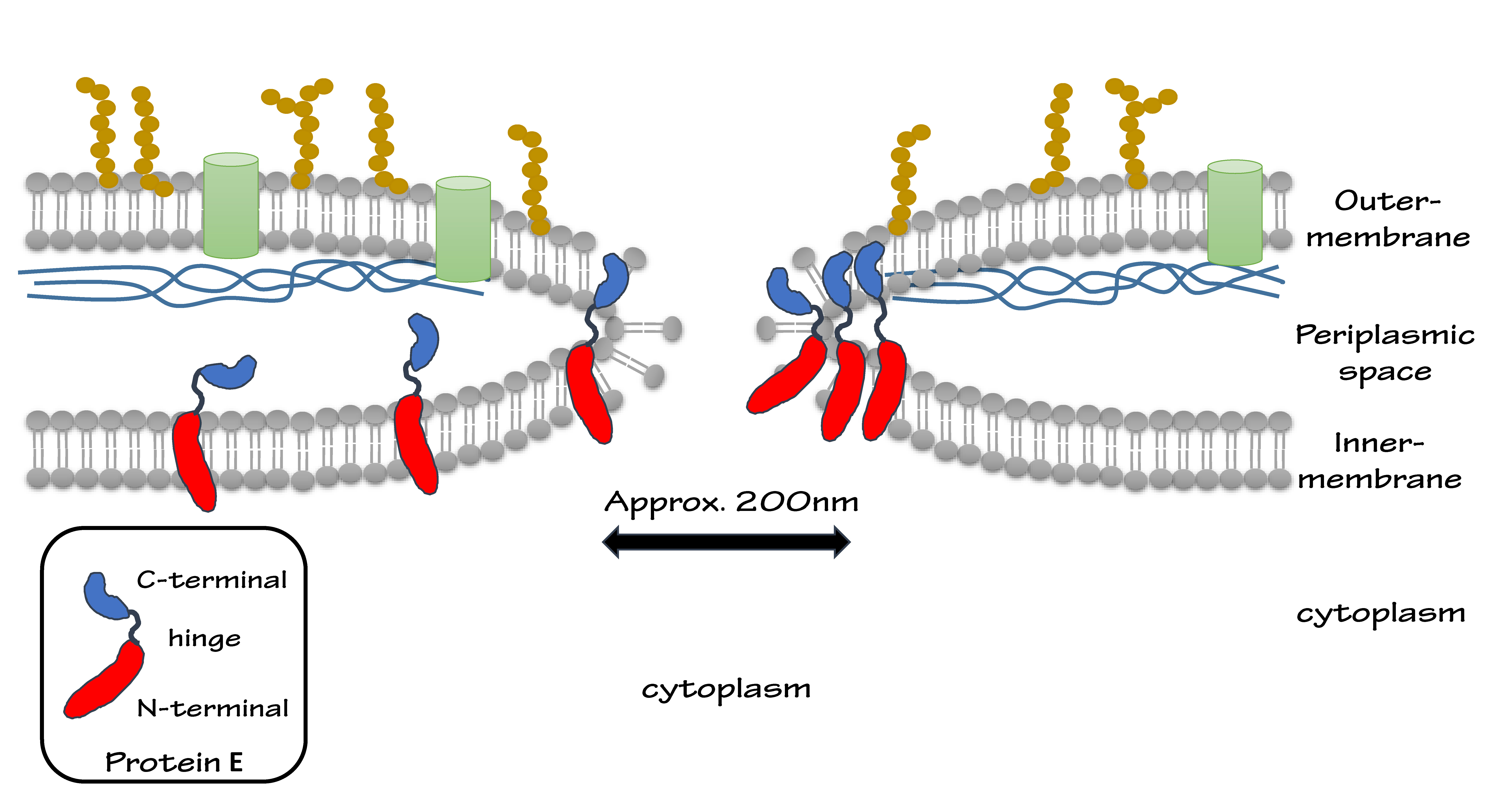
| - | We put Gram-negative bacteria lysis gene protein E under the control of pBAD promoter because of its short inducing time and L-arabinose concentration dependent trait. By making tunnels through inner and outer membrane of E.coli, we solve the conundrum that large molecules cannot enter the cells. At the same time, the periplasmic space remains intact, renders us the chance to utilize it as reaction | + | We put Gram-negative bacteria lysis gene '''protein E''' under the control of '''pBAD''' promoter because of its short inducing time and L-arabinose concentration dependent trait. By making tunnels through inner and outer membrane of ''E.coli'', we solve the conundrum that large molecules cannot enter the cells. At the same time, the periplasmic space remains intact, renders us the chance to utilize it as reaction site. |
| Line 17: | Line 17: | ||
[[File:Bacterialghost ppt.jpg|600px]] | [[File:Bacterialghost ppt.jpg|600px]] | ||
| - | (Protein E is a dual-domain protein cloned from bacteriophage ψX-174. The two domains of protein E help to fuse inner- and outer-membrane, keeping the periplasmic space intact.) | + | <small> |
| + | (Protein E is a '''dual-domain''' protein cloned from bacteriophage ψX-174. The two domains of protein E help to fuse inner- and outer-membrane, keeping the periplasmic space intact.) | ||
| + | </small> | ||
</center> | </center> | ||
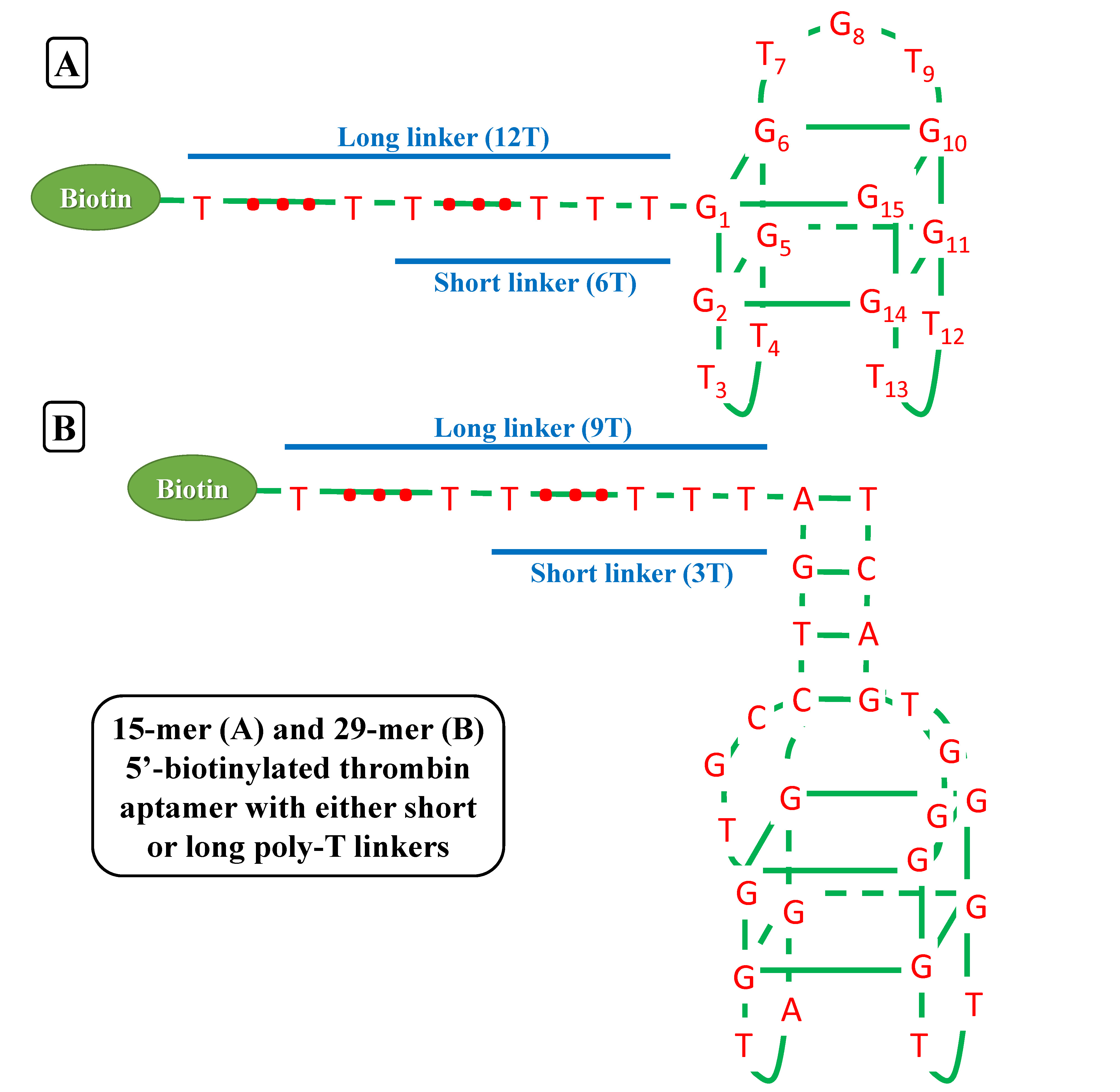
| - | We try to detect thrombin as our proof-of-concept experiment. There have been two well characterized aptamers, one is 15-mer and the other is 29-mer, all show very high and specific affinity to thrombin. We then connected the two aptamers with different length of poly T linkers, namely short and long linkers, to their 5’ end. At last, all aptamers were modified with 5’ biotin in order that they can bind to streptavidin. | + | We try to detect, purify and store '''thrombin''' as our proof-of-concept experiment. There have been two well characterized aptamers, one is '''15-mer''' and the other is '''29-mer''', all show very high and specific affinity to thrombin. We then connected the two aptamers with different length of '''poly-T''' linkers, namely short and long linkers, to their 5’ end. At last, all aptamers were modified with '''5’ biotin''' in order that they can bind to streptavidin. |
| - | [[ | + | [[File:15mer29merthrombinaptamer.jpg|400px|center]] |
| - | <html | + | <html><div class="clean"></div></html> |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
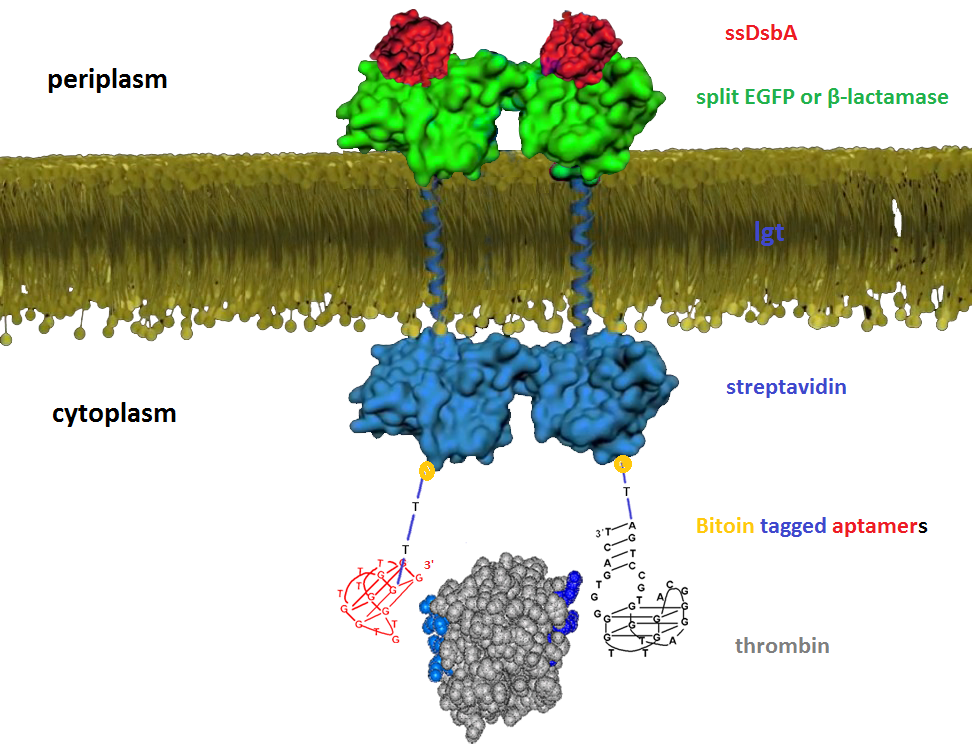
| + | We then built inner-membrane scaffolds consisted of three fusion parts. The split proteins were linked to the periplasmic ends of scaffolds and streptavidin to the cytoplasmic ends. We chose phosphatidylglycerol:: prolipoprotein diacylglyceryl transferase (Lgt) as our transmembrane protein because its function has been well identified by 2012 SJTU-BioX-Shanghai iGEM team. ssDsbA is the signal recognition particle (SRP)-dependent signaling sequence of DsbA. ssDsbA-tagged proteins are exported to the periplasm through the SRP pathway. With ssDsbA fused to the periplasmic terminal, fusion proteins with Lgt are expected to be anchored onto inner membrane of E.coli. | ||
<center> | <center> | ||
| - | [[File:ZJU-GhostSensor- | + | [[File:ZJU-GhostSensor-2.png|600px]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</center> | </center> | ||
| Line 59: | Line 39: | ||
<html> | <html> | ||
<div style="float:left;">Previous: | <div style="float:left;">Previous: | ||
| - | <a href="./Background">Ghost | + | <a href="./Background">Bacterial Ghost: Background</a> |
</div> | </div> | ||
<div style="float:right;">Next: | <div style="float:right;">Next: | ||
| - | <a href="./Results">Ghost | + | <a href="./Results">Bacterial Ghost: Results</a> |
</div> | </div> | ||
Latest revision as of 01:51, 29 October 2013
Bacterial Ghost: Design
We put Gram-negative bacteria lysis gene protein E under the control of pBAD promoter because of its short inducing time and L-arabinose concentration dependent trait. By making tunnels through inner and outer membrane of E.coli, we solve the conundrum that large molecules cannot enter the cells. At the same time, the periplasmic space remains intact, renders us the chance to utilize it as reaction site.
(Protein E is a dual-domain protein cloned from bacteriophage ψX-174. The two domains of protein E help to fuse inner- and outer-membrane, keeping the periplasmic space intact.)
We try to detect, purify and store thrombin as our proof-of-concept experiment. There have been two well characterized aptamers, one is 15-mer and the other is 29-mer, all show very high and specific affinity to thrombin. We then connected the two aptamers with different length of poly-T linkers, namely short and long linkers, to their 5’ end. At last, all aptamers were modified with 5’ biotin in order that they can bind to streptavidin.
We then built inner-membrane scaffolds consisted of three fusion parts. The split proteins were linked to the periplasmic ends of scaffolds and streptavidin to the cytoplasmic ends. We chose phosphatidylglycerol:: prolipoprotein diacylglyceryl transferase (Lgt) as our transmembrane protein because its function has been well identified by 2012 SJTU-BioX-Shanghai iGEM team. ssDsbA is the signal recognition particle (SRP)-dependent signaling sequence of DsbA. ssDsbA-tagged proteins are exported to the periplasm through the SRP pathway. With ssDsbA fused to the periplasmic terminal, fusion proteins with Lgt are expected to be anchored onto inner membrane of E.coli.
 "
"