Team:TU-Munich/Project/Localisation
From 2013.igem.org
m |
(→Verification of Localization – introducing a superior reporter protein) |
||
| (20 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
== Localization in ''Physcomitrella patens'' == | == Localization in ''Physcomitrella patens'' == | ||
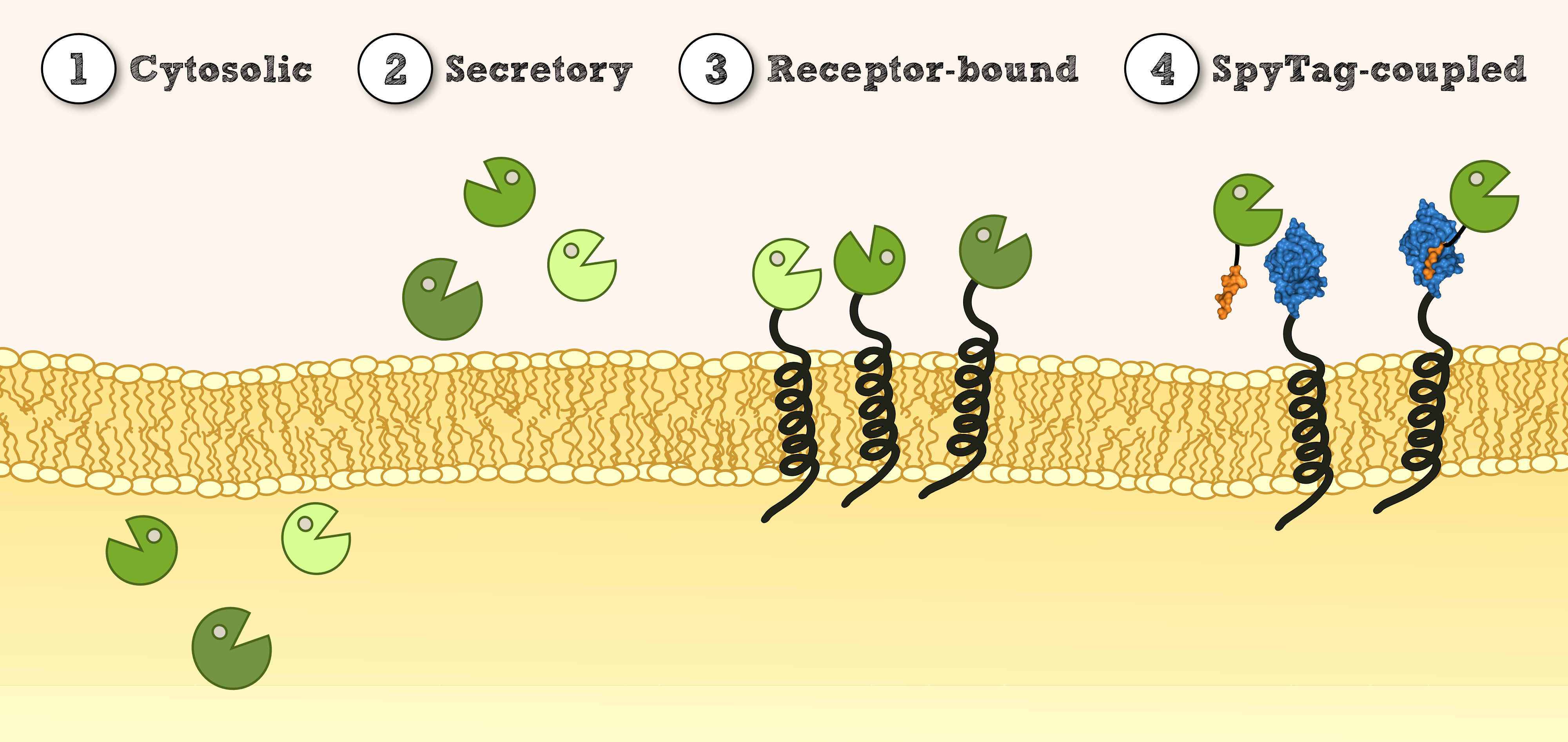
| - | [[File: | + | [[File:TUM13_Lokalisation_1.4.png|thumb|center|910px| '''Figure 1''': Overview of different possible localisations for effector proteins.]] |
In order to use ''P. patens'' as a chassis for Phytoremediaton, it is essential to be able to express effector proteins in different compartments. This includes '''cytoplasmatic expression''' of cytosolic effectors which degrade xenobiotics capable of crossing the cell membrane and which might depend on cofactors for degradation or conjugation. Secondly, there is the possibility to '''secrete effectors''' outside of the cell for an easy access to their respective target molecules. Other applications benefit from the expression of '''immobilized effectors''' on the inner or outer cellular membrane. This allows the creation of systems that do not release transgenic proteins into the environment and of systems that are able to internalize substances attached to recombinant binding proteins.<br> | In order to use ''P. patens'' as a chassis for Phytoremediaton, it is essential to be able to express effector proteins in different compartments. This includes '''cytoplasmatic expression''' of cytosolic effectors which degrade xenobiotics capable of crossing the cell membrane and which might depend on cofactors for degradation or conjugation. Secondly, there is the possibility to '''secrete effectors''' outside of the cell for an easy access to their respective target molecules. Other applications benefit from the expression of '''immobilized effectors''' on the inner or outer cellular membrane. This allows the creation of systems that do not release transgenic proteins into the environment and of systems that are able to internalize substances attached to recombinant binding proteins.<br> | ||
| - | '''Cytosolic''' expression of effector proteins was achieved by cloning the respective BioBrick downstream of the Actin_5 promoter. For the '''secretion''' of effectors, we compared several signal peptides with a prediction software and chose the signal peptide from the SERK receptor of ''Physcomitrella patens''. Additionally, we tested the signal peptides of a secreted antibody from mouse [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Gitzinger et al., 2009]] for comparison. For this purpose, the signal peptides were created as RFC 25 N-parts for the creation of fusion proteins with effectors available in RFC 25. Finally, the '''immobilization''' of recombinant effector proteins on a receptor which is functional in ''Physcomitrella patens'' was investigated by the construction of a synthetic receptor based on the SERK receptor. | + | '''Cytosolic''' expression of effector proteins was achieved by cloning the respective BioBrick downstream of the Actin_5 promoter. For the '''secretion''' of effectors, we compared several signal peptides with a prediction software and chose the signal peptide from the SERK receptor of ''Physcomitrella patens''. Additionally, we tested the signal peptides of a secreted antibody from mouse [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Gitzinger et al., 2009]] for comparison. For this purpose, the signal peptides were created as <nowiki>RFC 25</nowiki> N-parts for the creation of fusion proteins with effectors available in <nowiki>RFC 25</nowiki>. Finally, the '''immobilization''' of recombinant effector proteins on a receptor which is functional in ''Physcomitrella patens'' was investigated by the construction of a synthetic receptor based on the SERK receptor. |
=== Cytoplasmatic Expression === | === Cytoplasmatic Expression === | ||
| - | Cytoplasmatic expression of effector proteins is necessary | + | Cytoplasmatic expression of effector proteins is necessary for effectors that either depend on intracellular cofactors or for effectors with mechanisms linked to at least one exclusively intracellular component. For our project, this is the case for: |
| - | * Catechol-1,2-dioxygenase: | + | * Catechol-1,2-dioxygenase: Ferredoxins necessary to regenerate the oxygen-inactivated iron |
| - | * Glutathion-S-transferase: | + | * Glutathion-S-transferase: Glutathion necessary for coupling reactions |
=== Secretory Expression === | === Secretory Expression === | ||
| - | Effectors proteins | + | Effectors proteins containing '''disulfide bonds''' have to be secreted, as the reducing milieu of the cytosolic compartment does not allow formation of disulfid bonds. For secretion, the protein has to be fused to a '''signal peptide''' for the translocation into the endoplasmatic reticulum. Therefore we created two BioBricks encoding a signal sequences for endoplasmatic reticular translocation: The '''SERK signal peptide''' ([http://parts.igem.org/Part:BBa_K1159303 BBa_K1159303]) of the Somatic Embryogenesis Receptor Kinase from ''Physcomitrella patens'' (see [https://2013.igem.org/Team:TU-Munich/Modeling/Protein_Predictions#Prediction_of_Signal_Peptides Prediction of Signal Peptides] for more details) and the '''IgKappa signal peptide''' of the Ig Kappa chain from ''Mus musculus'' ([http://parts.igem.org/Part:BBa_K1159304 BBa_K1159304]) [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]]. Our following effector needs to be secreted: |
| - | * Laccase from ''Bacillus pumilus'': | + | * Laccase from ''Bacillus pumilus'': 1 disulfide bond |
=== Membrane-bound Expression === | === Membrane-bound Expression === | ||
| Line 30: | Line 30: | ||
[[File:TUM13_SERK-Receptor.png|thumb|right|300px| '''Figure 2''': Schematic design of an effector immobilized on a transmembrane domain.]] | [[File:TUM13_SERK-Receptor.png|thumb|right|300px| '''Figure 2''': Schematic design of an effector immobilized on a transmembrane domain.]] | ||
| - | For '''substance binding proteins''' or as | + | For '''substance binding proteins''' or as an '''alternative to secretion''' for degrading enzymes, it is convenient to anchor effectors on the plant´s surface inside its cytoplasmatic membrane. |
| - | For this purpose, we | + | For this purpose, we added a '''signal for secretion''' at the N-terminus of our effector protein as well as a downstream '''transmembrane domain''' from ''Physcomitrella patens'' ([http://parts.igem.org/Part:BBa_K1159315 BBa_K1159315]) for membrane localization. Furthermore, we fused GFP ([http://parts.igem.org/Part:BBa_K1159311 BBa_K1159311]) to the intracellular site of the transmembrane region for the verification of membrane localization via fluorescence microscopy. We set a '''linker''' containing ''Strep''-tag II and a TEV cleavage site in between the effector protein and the transmembrane region. This allows us to characterize the membrane-bound effectors by incubating the plant with TEV Protease and thus releasing the respective effectors. Hence, purification via the ''Strep''-tag II and successive characterization of the effector is possible. |
For more details on the design process of the transmembrane region, see [https://2013.igem.org/Team:TU-Munich/Modeling/Protein_Predictions Protein Predictions]. | For more details on the design process of the transmembrane region, see [https://2013.igem.org/Team:TU-Munich/Modeling/Protein_Predictions Protein Predictions]. | ||
| Line 39: | Line 39: | ||
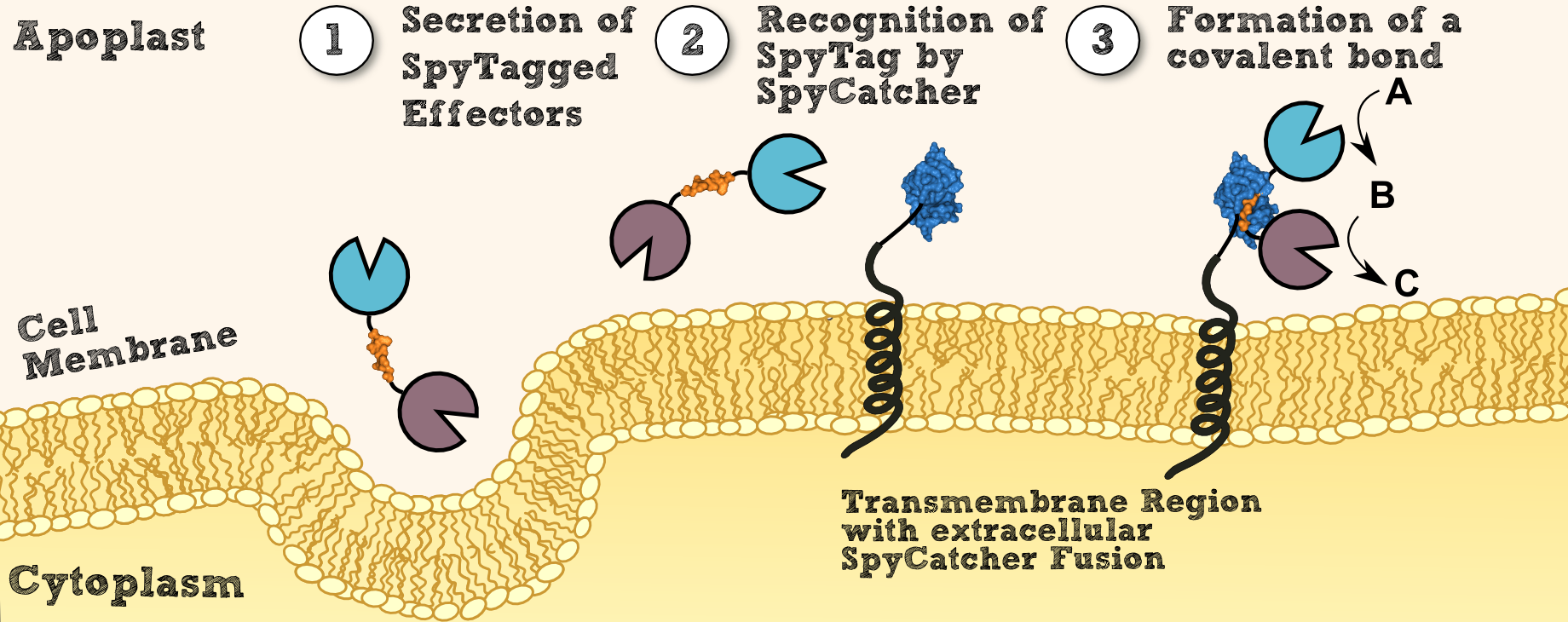
[[File:SpyCatcherSpyTag.png|thumb|center|910px|'''Figure 3''': Steps of the localization process via SpyTag and SpyCatcher.]] | [[File:SpyCatcherSpyTag.png|thumb|center|910px|'''Figure 3''': Steps of the localization process via SpyTag and SpyCatcher.]] | ||
| - | To create transgenic moss able to degrade customized combinations of xenobiotics targeted by different effector molecules, we introduced the SpyCatcher/SpyTag system into our project [[http://www.pnas.org/content/109/12/E690 Bijan Zakeri et al., 2012]]. This system was created for posttranslational protein fusion based on a covalent bond which is formed between the side chains of residues of SypCatcher and SpyTag. SpyCatcher and SpyTag | + | To create transgenic moss that is able to degrade customized combinations of xenobiotics targeted by different effector molecules, we introduced the SpyCatcher/SpyTag system into our project [[http://www.pnas.org/content/109/12/E690 Bijan Zakeri et al., 2012]]. This system was created for '''posttranslational protein fusion''' based on a covalent bond which is formed between the side chains of residues of SypCatcher and SpyTag. SpyCatcher and SpyTag were created by splitting and engineering the FbaB domain from ''Streptococcus pyogenes''. The splitted parts recognize each other and form a '''covalent isopeptide bridge''', like in their natural non-splitted form. |
| - | Recombinant effector proteins fused to the SpyTag can thus be '''expressed separately''' (e.g. at a different intensity, via a secretory pathway) from receptors fused to the SypCatcher. | + | Recombinant effector proteins fused to the SpyTag can thus be '''expressed separately''' (e.g. at a different intensity, via a secretory pathway) from receptors fused to the SypCatcher. This enables us to control the SERK-receptor´s expression through a strong promoter while individually adjusting the expression of different effector proteins according to their respectively desired concentration (see Fig. 3.2). |
| - | Another application lies in the fusion of multimeric effector proteins to receptors, where the fusion of all individual subunits to the receptor is not a viable option due to '''steric reasons'''. The SpyCatcher/SpyTag system bypasses this problem as it allows the multimeric protein to assemble into its active form before its immobilization on the exterior membrane by a respective receptor | + | Another application lies in the '''fusion of multimeric effector proteins to receptors''', where the fusion of all individual subunits to the receptor is not a viable option due to '''steric reasons'''. The SpyCatcher/SpyTag system bypasses this problem, as it allows the multimeric protein to assemble into its active form before its immobilization on the exterior membrane by a respective receptor (see Fig. 3.3). |
But the biggest advantage of the system is that it allows you '''non-canonical post-translational protein fusions'''. With this system '''you can fuse e.g. the C-terminus of a protein not only with the N-terminus of another protein but also the C-terminus.''' The reason is that the formation of the covalent bond is not a canonical peptide bond but a isopeptide bond between the side residues of SpyCatcher and SpyTag. | But the biggest advantage of the system is that it allows you '''non-canonical post-translational protein fusions'''. With this system '''you can fuse e.g. the C-terminus of a protein not only with the N-terminus of another protein but also the C-terminus.''' The reason is that the formation of the covalent bond is not a canonical peptide bond but a isopeptide bond between the side residues of SpyCatcher and SpyTag. | ||
| - | + | It also allows you to create membrane-bound enzyme assembly line for two-step catalyzed reactions by fusing the two involved enzymes to the both termini of SpyTag. | |
| - | [[File:TUM13_Annimated_test.gif|thumb|right|300px| '''Figure | + | [[File:SpyCatcherSpyTag_assembly_line.png|thumb|center|910px|'''Figure 4''': Steps of the localization process of a fusion protein consisting two enzymes via SpyCatcher/SpyTag system, resulting in a branch structure.]] |
| - | To analyse the proper localization of our moss constructs we created a BioBrick from the '''NanoLuc''' Luciferase by Promega. NanoLuc Luciferase can be up to 240x brighter than conventional firefly luciferase and is at least two times smaller than other luciferases. This makes the NanoLuc Luciferase an '''ideal reporter protein''' which can not only be used to verify proper localisation but can also be employed whenever a low detection threshold is needed. | + | |
| + | ==Verification of Localization – introducing a superior reporter protein== | ||
| + | |||
| + | [[File:TUM13_Annimated_test.gif|thumb|right|300px| '''Figure 5:''' Homologous structure (3ppt_A) of the NanoLuc Luciferase]] | ||
| + | To analyse the proper localization of our moss constructs we created a [http://parts.igem.org/wiki/index.php/Part:BBa_K1159001 BioBrick BBa_K1159001] from the '''NanoLuc''' Luciferase by Promega. NanoLuc Luciferase can be up to 240x brighter than conventional firefly luciferase and is at least two times smaller than other luciferases. This makes the NanoLuc Luciferase an '''ideal reporter protein''' which can not only be used to verify proper localisation, but can also be employed whenever a low detection threshold is needed. | ||
==References:== | ==References:== | ||
Latest revision as of 02:58, 29 October 2013
Localization in Physcomitrella patens
In order to use P. patens as a chassis for Phytoremediaton, it is essential to be able to express effector proteins in different compartments. This includes cytoplasmatic expression of cytosolic effectors which degrade xenobiotics capable of crossing the cell membrane and which might depend on cofactors for degradation or conjugation. Secondly, there is the possibility to secrete effectors outside of the cell for an easy access to their respective target molecules. Other applications benefit from the expression of immobilized effectors on the inner or outer cellular membrane. This allows the creation of systems that do not release transgenic proteins into the environment and of systems that are able to internalize substances attached to recombinant binding proteins.
Cytosolic expression of effector proteins was achieved by cloning the respective BioBrick downstream of the Actin_5 promoter. For the secretion of effectors, we compared several signal peptides with a prediction software and chose the signal peptide from the SERK receptor of Physcomitrella patens. Additionally, we tested the signal peptides of a secreted antibody from mouse http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Gitzinger et al., 2009 for comparison. For this purpose, the signal peptides were created as RFC 25 N-parts for the creation of fusion proteins with effectors available in RFC 25. Finally, the immobilization of recombinant effector proteins on a receptor which is functional in Physcomitrella patens was investigated by the construction of a synthetic receptor based on the SERK receptor.
Cytoplasmatic Expression
Cytoplasmatic expression of effector proteins is necessary for effectors that either depend on intracellular cofactors or for effectors with mechanisms linked to at least one exclusively intracellular component. For our project, this is the case for:
- Catechol-1,2-dioxygenase: Ferredoxins necessary to regenerate the oxygen-inactivated iron
- Glutathion-S-transferase: Glutathion necessary for coupling reactions
Secretory Expression
Effectors proteins containing disulfide bonds have to be secreted, as the reducing milieu of the cytosolic compartment does not allow formation of disulfid bonds. For secretion, the protein has to be fused to a signal peptide for the translocation into the endoplasmatic reticulum. Therefore we created two BioBricks encoding a signal sequences for endoplasmatic reticular translocation: The SERK signal peptide ([http://parts.igem.org/Part:BBa_K1159303 BBa_K1159303]) of the Somatic Embryogenesis Receptor Kinase from Physcomitrella patens (see Prediction of Signal Peptides for more details) and the IgKappa signal peptide of the Ig Kappa chain from Mus musculus ([http://parts.igem.org/Part:BBa_K1159304 BBa_K1159304]) http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009. Our following effector needs to be secreted:
- Laccase from Bacillus pumilus: 1 disulfide bond
Membrane-bound Expression
For substance binding proteins or as an alternative to secretion for degrading enzymes, it is convenient to anchor effectors on the plant´s surface inside its cytoplasmatic membrane.
For this purpose, we added a signal for secretion at the N-terminus of our effector protein as well as a downstream transmembrane domain from Physcomitrella patens ([http://parts.igem.org/Part:BBa_K1159315 BBa_K1159315]) for membrane localization. Furthermore, we fused GFP ([http://parts.igem.org/Part:BBa_K1159311 BBa_K1159311]) to the intracellular site of the transmembrane region for the verification of membrane localization via fluorescence microscopy. We set a linker containing Strep-tag II and a TEV cleavage site in between the effector protein and the transmembrane region. This allows us to characterize the membrane-bound effectors by incubating the plant with TEV Protease and thus releasing the respective effectors. Hence, purification via the Strep-tag II and successive characterization of the effector is possible.
For more details on the design process of the transmembrane region, see Protein Predictions.
Modularization: Post-translational fusion using SpyTag & SpyCatcher
To create transgenic moss that is able to degrade customized combinations of xenobiotics targeted by different effector molecules, we introduced the SpyCatcher/SpyTag system into our project http://www.pnas.org/content/109/12/E690 Bijan Zakeri et al., 2012. This system was created for posttranslational protein fusion based on a covalent bond which is formed between the side chains of residues of SypCatcher and SpyTag. SpyCatcher and SpyTag were created by splitting and engineering the FbaB domain from Streptococcus pyogenes. The splitted parts recognize each other and form a covalent isopeptide bridge, like in their natural non-splitted form.
Recombinant effector proteins fused to the SpyTag can thus be expressed separately (e.g. at a different intensity, via a secretory pathway) from receptors fused to the SypCatcher. This enables us to control the SERK-receptor´s expression through a strong promoter while individually adjusting the expression of different effector proteins according to their respectively desired concentration (see Fig. 3.2).
Another application lies in the fusion of multimeric effector proteins to receptors, where the fusion of all individual subunits to the receptor is not a viable option due to steric reasons. The SpyCatcher/SpyTag system bypasses this problem, as it allows the multimeric protein to assemble into its active form before its immobilization on the exterior membrane by a respective receptor (see Fig. 3.3).
But the biggest advantage of the system is that it allows you non-canonical post-translational protein fusions. With this system you can fuse e.g. the C-terminus of a protein not only with the N-terminus of another protein but also the C-terminus. The reason is that the formation of the covalent bond is not a canonical peptide bond but a isopeptide bond between the side residues of SpyCatcher and SpyTag.
It also allows you to create membrane-bound enzyme assembly line for two-step catalyzed reactions by fusing the two involved enzymes to the both termini of SpyTag.
Verification of Localization – introducing a superior reporter protein
To analyse the proper localization of our moss constructs we created a [http://parts.igem.org/wiki/index.php/Part:BBa_K1159001 BioBrick BBa_K1159001] from the NanoLuc Luciferase by Promega. NanoLuc Luciferase can be up to 240x brighter than conventional firefly luciferase and is at least two times smaller than other luciferases. This makes the NanoLuc Luciferase an ideal reporter protein which can not only be used to verify proper localisation, but can also be employed whenever a low detection threshold is needed.
References:
http://www.plantphysiol.org/content/127/4/1430 Schaefer and Zryd, 2001 Schaefer, D.G. and Zrÿd, J. (2001). The Moss Physcomitrella patens, Now and Then. Plant Physiology, 127(4):1430-1438.
http://www.ncbi.nlm.nih.gov/pubmed/22366317 Zakeri et al., 2012 Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 20;109(12)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3315829/ Hagan et al., 2010 Hagan RM, Björnsson R, McMahon SA, Schomburg B, Braithwaite V, Bühl M, Naismith JH, Schwarz-Linek U. (2010). NMR spectroscopic and theoretical analysis of a spontaneously formed Lys-Asp isopeptide bond. Angew Chem Int Ed Engl. 2;49(45)
http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Gitzinger M, Parsons J, Reski R, Fussenegger M (2009). Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries. Plant Biotechnol J. 7(1):73-86.
 "
"





AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351