Team:TU-Munich/Results/KillSwitch
From 2013.igem.org
(→Transformation of the large DNA constructs) |
(→Photosensitivity) |
||
| (37 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
==Kill Switch== | ==Kill Switch== | ||
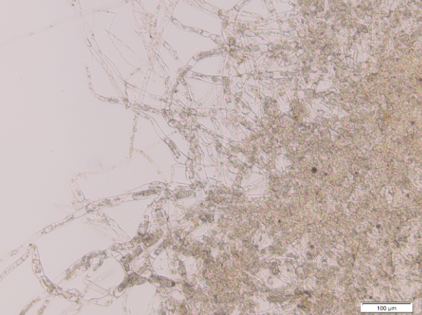
| + | [[File:TUM13_Moostot_(2).png|thumb|left|300px| '''Figure 1''': Dead Kill Switch moss protonema]] | ||
| - | The Kill Switch mechanism is the most complex and ambitious aspect of our project on the protein level | + | The Kill Switch mechanism is the most complex and ambitious aspect of our project on the protein level. For details on its design and function see [https://2013.igem.org/Team:TU-Munich/Project/Killswitch here]. We were very excited to start experimenting with the killswitch moss. |
| - | + | However, when we opened the redlight filter foil, we found both the PIF3 and the PIF6 version of the allegedly transgenic moss lines were dead. There is a variety of possible reasons for this outcome that need to be discussed in order to improve our approach and produce live Kill Switch moss lines. | |
==Transformation of the large DNA constructs== | ==Transformation of the large DNA constructs== | ||
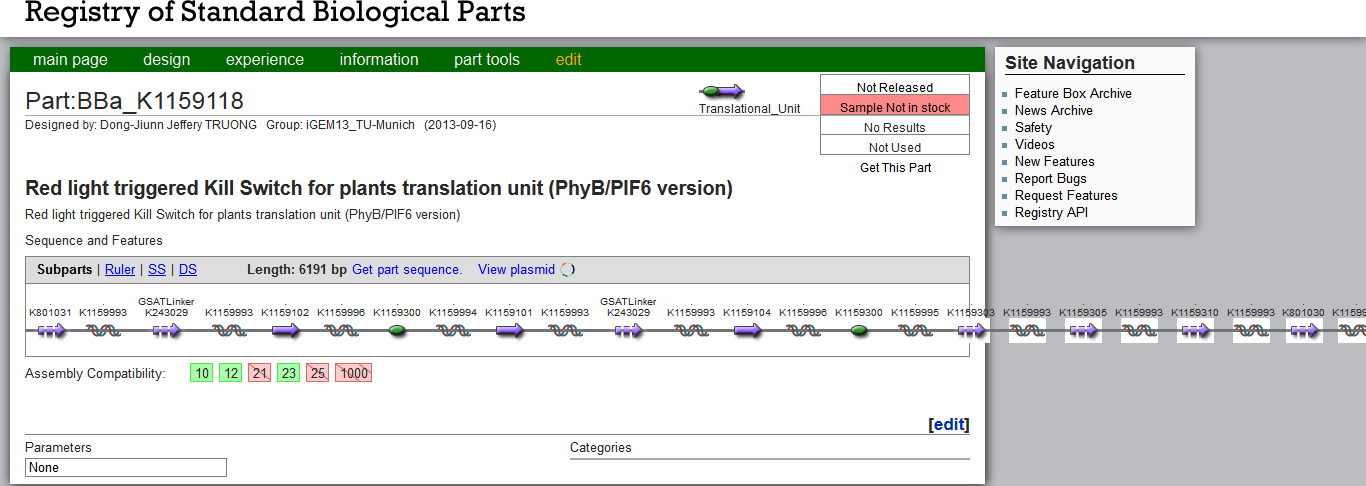
| - | For the PEG-mediated moss [https://2013.igem.org/Team:TU-Munich/Notebook/Methods#Transformation transformation], we used plasmids linearized with EcoRI | + | The Kill Switch DNA constructs are very large and even exceed the Part Registry´s frame (see Figure 2). For the PEG-mediated moss [https://2013.igem.org/Team:TU-Munich/Notebook/Methods#Transformation transformation], we used plasmids linearized with EcoRI, but contrary to the targeted gene transfer described by [http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2003], our constructs contained no flanking homologuos regions. Our random integration transformation worked successfully for our other contructs, whereas the large Kill Switch construct might need further inquiry for a suitable integration site. Due to its size, the construct might be less stable and require sensitive handling. These factors could have lead to a failed transformation, in which case the moss would have died through the G418 selection process. |
| + | |||
| + | [[File:TUM13_PartsRegistryKillSwitch.png|thumb|center|650px| '''Figure 2''': [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159118 Registry entry] of the PhyB/PIF6 version of the Kill Switch, BBa_K1159118]] | ||
We weren´t yet able to express the Kill Switch in ''E.coli'', which is most likely due to codon usage discrepancy. While ''Physcomitrella patens'' is not very dicriminating regarding codon usage, the function and expression of proteins designed for ''Physco'' is not guaranteed to work in ''E.coli''. | We weren´t yet able to express the Kill Switch in ''E.coli'', which is most likely due to codon usage discrepancy. While ''Physcomitrella patens'' is not very dicriminating regarding codon usage, the function and expression of proteins designed for ''Physco'' is not guaranteed to work in ''E.coli''. | ||
| Line 20: | Line 23: | ||
==Level of promoter activity== | ==Level of promoter activity== | ||
| - | [[File:PBI_376_f1.gif|thumb|right| | + | [[File:PBI_376_f1.gif|thumb|right|300px| '''Figure 4''': Comparative expression performance of different constitutive mammalian and plant promoters in Physcomitrella patens, [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]]]] |
| - | + | We used the Actin 5 promotor from ''Physcomitrella patens'' [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159306 BBa_K1159306], which is the strongest known Physco promotor (see Figure 3). The very high intensity of expression might lead to problems concerning the vitality of the transgenic moss. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| + | Because the Kill Switch encodes for the very effective thermonuclease, overexpression might lead to loss of vitality in the moss regardless of its fixation to the cell membrane. Equipping the Kill Switch with a promotor for a lower level of expression, such as the CMV promotor or the SV40 promotor, could maybe lead to a higher survival rate of successfully transformed moss. | ||
| + | ==Photosensitivity== | ||
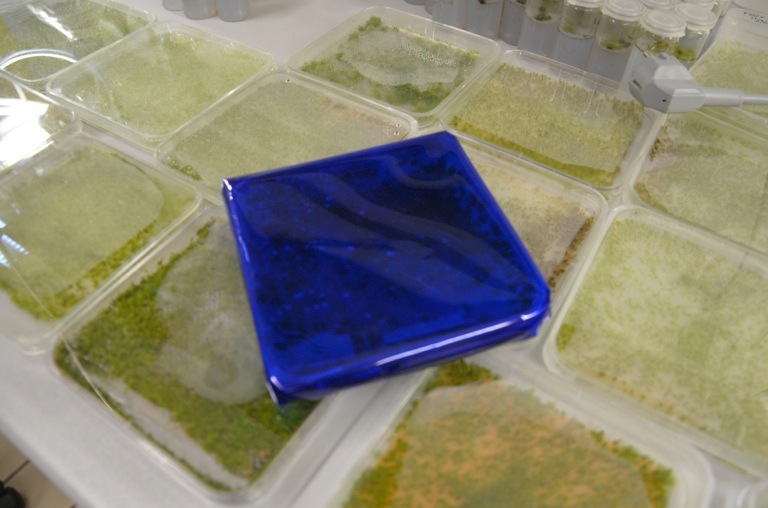
| + | [[File:TUM13_Moosfolie.jpg|thumb|right|250px| '''Figure 4''': Photosensitive moss protected by foil wrapping]] | ||
| + | The programmed cell death triggered through the Kill Switch is induced through the red light fraction of the spectrum of visible light. It is therefore crucial to shield transformed moss plants from light! We used a layer combination green and blue foil that was wrapped arouns the protoplast 6-wells right after transformation, (see Figure 4). | ||
| + | [[File:TUM13_Foto_Labpics2.jpg|thumb|right|250px| '''Figure 5''': Working with the photosensitive protoplasts]] | ||
| + | To start the selection process, the still vital regenerated moss cells were transfered onto G418 agar plates at night, lighting the workspace with a lamp wrapped up in the previously described foils, (see Figure 5). | ||
| + | Since the moss survived until this point, we can conclude that the light the foil allows to pass is sufficient for the moss to grow. Its early death is therefore not due to insufficient light supply. Given the precautions we met in the handling process, accidental contact with unfiltered light can be ruled out as a reason. | ||
| + | However, it is not guaranteed that the foil layers are sufficient to protect the moss from red light and of course we could not yet determine which wavelenghts define the limits of the spectrum the moss can tolerate and which intensity of light inside the spectrum of interest is needed to trigger the TEV protease fusion. | ||
==References== | ==References== | ||
| Line 40: | Line 44: | ||
[[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]] Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries, Plant Biotechnol J. 2009 Feb;7(2):210 | [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]] Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries, Plant Biotechnol J. 2009 Feb;7(2):210 | ||
| - | [[http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., | + | [[http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2004]] An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, ''Physcomitrella patens'', Curr Genet (2004) 44: 339–347 |
<div class="visualClear"></div> | <div class="visualClear"></div> | ||
Latest revision as of 03:26, 29 October 2013
Kill Switch
The Kill Switch mechanism is the most complex and ambitious aspect of our project on the protein level. For details on its design and function see here. We were very excited to start experimenting with the killswitch moss.
However, when we opened the redlight filter foil, we found both the PIF3 and the PIF6 version of the allegedly transgenic moss lines were dead. There is a variety of possible reasons for this outcome that need to be discussed in order to improve our approach and produce live Kill Switch moss lines.
Transformation of the large DNA constructs
The Kill Switch DNA constructs are very large and even exceed the Part Registry´s frame (see Figure 2). For the PEG-mediated moss transformation, we used plasmids linearized with EcoRI, but contrary to the targeted gene transfer described by [http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2003], our constructs contained no flanking homologuos regions. Our random integration transformation worked successfully for our other contructs, whereas the large Kill Switch construct might need further inquiry for a suitable integration site. Due to its size, the construct might be less stable and require sensitive handling. These factors could have lead to a failed transformation, in which case the moss would have died through the G418 selection process.
We weren´t yet able to express the Kill Switch in E.coli, which is most likely due to codon usage discrepancy. While Physcomitrella patens is not very dicriminating regarding codon usage, the function and expression of proteins designed for Physco is not guaranteed to work in E.coli.
Level of promoter activity
We used the Actin 5 promotor from Physcomitrella patens [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159306 BBa_K1159306], which is the strongest known Physco promotor (see Figure 3). The very high intensity of expression might lead to problems concerning the vitality of the transgenic moss.
Because the Kill Switch encodes for the very effective thermonuclease, overexpression might lead to loss of vitality in the moss regardless of its fixation to the cell membrane. Equipping the Kill Switch with a promotor for a lower level of expression, such as the CMV promotor or the SV40 promotor, could maybe lead to a higher survival rate of successfully transformed moss.
Photosensitivity
The programmed cell death triggered through the Kill Switch is induced through the red light fraction of the spectrum of visible light. It is therefore crucial to shield transformed moss plants from light! We used a layer combination green and blue foil that was wrapped arouns the protoplast 6-wells right after transformation, (see Figure 4).
To start the selection process, the still vital regenerated moss cells were transfered onto G418 agar plates at night, lighting the workspace with a lamp wrapped up in the previously described foils, (see Figure 5).
Since the moss survived until this point, we can conclude that the light the foil allows to pass is sufficient for the moss to grow. Its early death is therefore not due to insufficient light supply. Given the precautions we met in the handling process, accidental contact with unfiltered light can be ruled out as a reason.
However, it is not guaranteed that the foil layers are sufficient to protect the moss from red light and of course we could not yet determine which wavelenghts define the limits of the spectrum the moss can tolerate and which intensity of light inside the spectrum of interest is needed to trigger the TEV protease fusion.
References
http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries, Plant Biotechnol J. 2009 Feb;7(2):210
http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2004 An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens, Curr Genet (2004) 44: 339–347
 "
"




AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351