Team:BYU Provo/Notebook
From 2013.igem.org
(Difference between revisions)
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{TeamBYUProvo}} | {{TeamBYUProvo}} | ||
| - | + | [[File:BYUNotebook.JPG|965px|center|link=]] | |
| - | + | =='''Phage Team'''== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| width="100%" style="text-align: top;" | {| width="100%" style="text-align: top;" | ||
| Line 21: | Line 10: | ||
| style="width: 22%; background-color: transparent;"| | | style="width: 22%; background-color: transparent;"| | ||
| - | [[File:BYULPProjectPic.JPG|220px|center]] | + | [[File:BYULPProjectPic.JPG|220px|center|link=]] |
| style="width: 58%; background-color: transparent;"| | | style="width: 58%; background-color: transparent;"| | ||
| Line 31: | Line 20: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | : '''Bryan Merrill and Keltzie Westra''' |
| + | |||
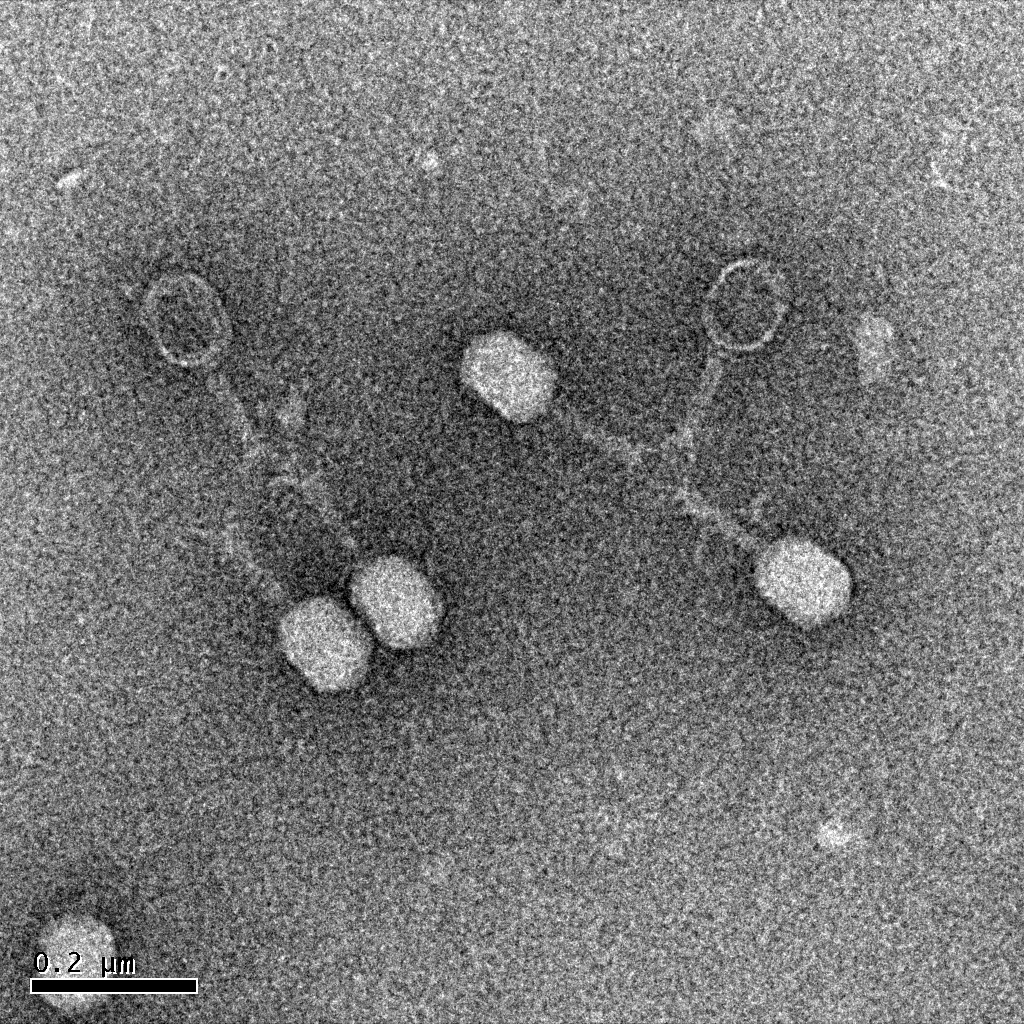
| + | : The focus of the Large Phage Team is to make a cross section of phage sizes through the mutagenesis of T4 bacteriophage. Our main area of research is in making T4's already large capsid even larger, while maintaining phenotypic stability. This was done by taking the isolated large phage and observing there later generations to ensure that their phenotype is inherited. | ||
</font> | </font> | ||
| Line 57: | Line 48: | ||
|- | |- | ||
| - | | style="width: 22%; background-color: | + | | style="width: 22%; background-color: transparent;"| |
| - | + | [[File:S4.JPG|220px|center|link=]] | |
| style="width: 53%; background-color: transparent;"| | | style="width: 53%; background-color: transparent;"| | ||
| Line 68: | Line 59: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : ''' | + | : '''Xiuqi (Jade) Li and LJ Perry''' |
| - | + | ||
| - | + | ||
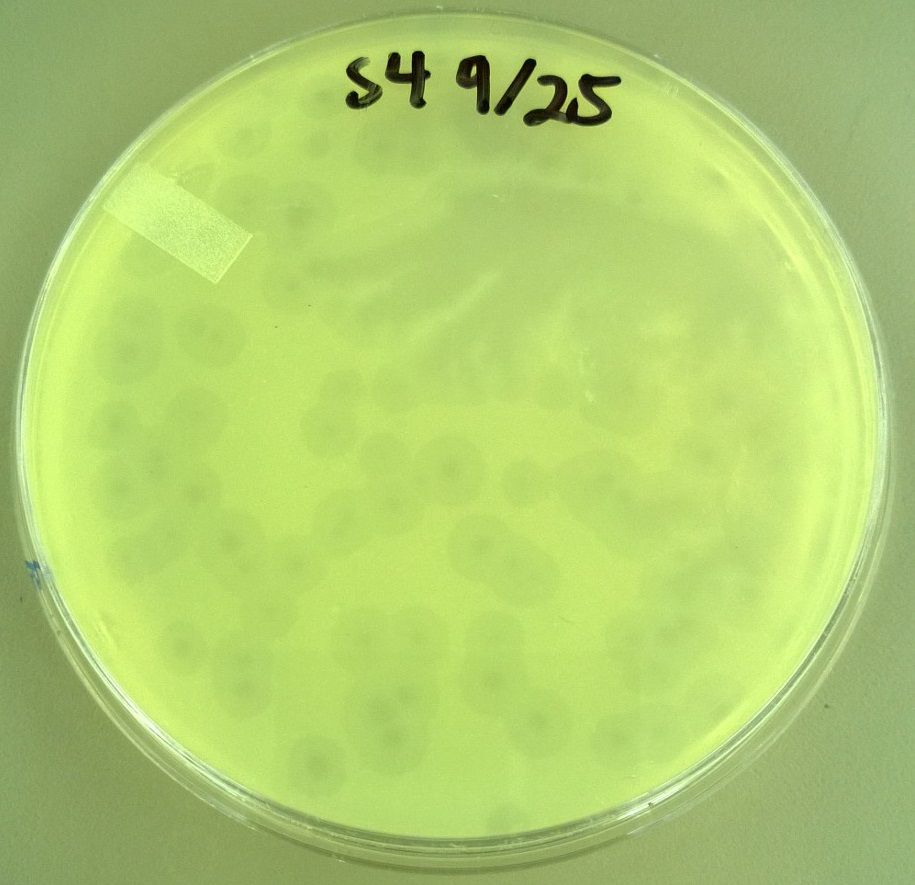
| - | : We are creating a library of different size T7 bacteriophage capsids. This is accomplished by mutating the phage with a base analog and then isolating the smaller and bigger T7 phages. We are also | + | : We are creating a library of different size T7 bacteriophage capsids. This is accomplished by mutating the phage with a base analog and then isolating the smaller and bigger T7 phages. We are also establishing the relationship between the capsid size and the plaque size of certain phages. |
</font> | </font> | ||
| Line 100: | Line 89: | ||
| style="width: 22%; background-color: transparent;"| | | style="width: 22%; background-color: transparent;"| | ||
| - | [[File:Cesiumchloridegradient.jpg|220px|center]] | + | [[File:Cesiumchloridegradient.jpg|220px|center|link=]] |
| style="width: 53%; background-color: transparent;"| | | style="width: 53%; background-color: transparent;"| | ||
| Line 110: | Line 99: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : As members of the | + | : '''Amber Brown, Arick Christopher, and Darren Lasko''' |
| + | |||
| + | : As members of the Phage Purification Team, we focus on the purification of bacteriophages and the preparation of ghost capsids. We have worked with several different methods of preparation and can manipulate the procedures to accommodate the type of phage and the capsid size of the mutants. | ||
</font> | </font> | ||
| Line 138: | Line 129: | ||
<br> | <br> | ||
| - | ==Cholera Team== | + | =='''Cholera Team'''== |
{| width="100%" style="text-align: top;" | {| width="100%" style="text-align: top;" | ||
|- valign="center" | |- valign="center" | ||
| - | | style="width: 22%; background-color: | + | | style="width: 22%; background-color: transparent;"| |
| - | + | [[File:BYUCCCC.jpg|220px|center|link=]] | |
| style="width: 58%; background-color: transparent;"| | | style="width: 58%; background-color: transparent;"| | ||
| Line 155: | Line 146: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | : '''Clarice Harrison, Kendall Kiser, and Kelton Peck''' |
| + | |||
| + | : Our focus is two-fold: 1) develop a system in ''E.coli'' to detect the presence ''V. cholerae'', and 2) integrate ''E.coli'' bacteriophage lambda into a biofilm-degrading machine that is moderated by our detection system. Upon discovering experimentally that bacteriophage lambda can naturally sense cholera, our research efforts have centered on synthetically improving this pathway. | ||
</font> | </font> | ||
| Line 181: | Line 174: | ||
|- | |- | ||
| - | | style="width: 22%; background-color: | + | | style="width: 22%; background-color: transparent;"| |
| - | + | [[File:BYU2013-CholeraGroup1.jpg|220px|center|link=]] | |
| style="width: 53%; background-color: transparent;"| | | style="width: 53%; background-color: transparent;"| | ||
| Line 193: | Line 186: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | : '''Nathan Sabin and Michael Schellhous''' |
| + | |||
| + | : On the Cholera-Enzyme team our focus is researching enzymes that will degrade the ''V. cholerae'' biofilm. Our goal is to find enzymes that have potential to degrade the biofilm, isolate them, clone them into ''E. coli'', and design an assay to test their biofilm-degrading ability. | ||
</font> | </font> | ||
Latest revision as of 03:49, 28 September 2013
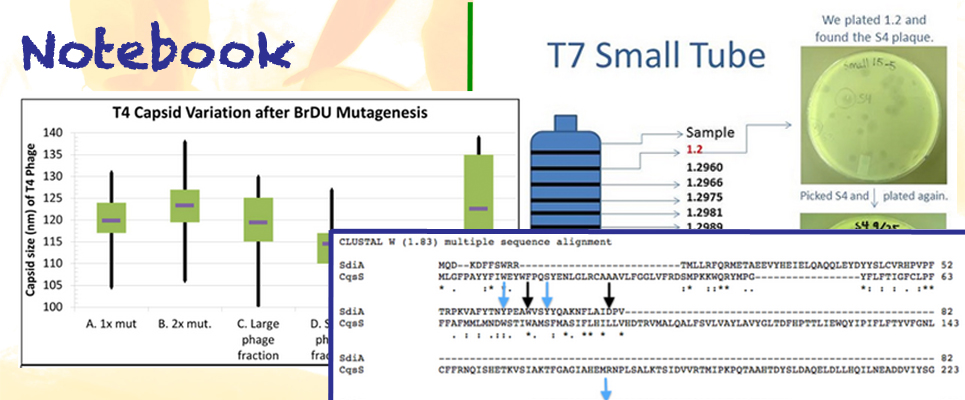
Phage Team
 |
|
|
 |
|
|
 |
|
|
Cholera Team
 |
|
|
 |
|
|
 "
"