Team:Paris Saclay/Notebook/August/21
From 2013.igem.org
(Difference between revisions)
(→1 - Electrophoresis of the digestion of BBa_J04450 by EcoRI/PstI) |
|||
| (20 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
==='''A - Aerobic/Anaerobic regulation system'''=== | ==='''A - Aerobic/Anaerobic regulation system'''=== | ||
| - | ====''' | + | ===='''Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006'''==== |
| - | + | ====1 - Colony PCR of NarK, NarG or NirB with RBS-LacZ-Term or RBS-AmilCP-Term in pSB1C3 in DH5α==== | |
| - | + | ||
| - | + | Damir, Xiaojing | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | {| | |
| - | + | | style="border:1px solid black;padding:5px;background-color:#DE;" | | |
| - | + | The transformation was good. We will do a Colony PCR. | |
| - | + | |} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Colonies repiquée dans 10µL d'eau. | ||
| + | We do 8 PCR for each ligation. | ||
| - | + | Used quantities : | |
| + | * Mix A : | ||
| + | ** Buffer Dream Taq : 250µL | ||
| + | ** dNTP : 50µL | ||
| + | ** Oligo 44 : 50µL | ||
| + | ** Dream Taq : 20µL | ||
| + | ** H2O : 1.88mL | ||
| + | *** NirB with RBS-LacZ-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 45+2µL of DNA | ||
| + | *** NirB with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 45 +2µL of DNA | ||
| + | *** NarG with RBS-LacZ-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 41+2µL of DNA | ||
| + | *** NarG with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 41 +2µL of DNA | ||
| + | *** NarK with RBS-LacZ-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 47+2µL of DNA | ||
| + | *** NarK with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 45µL of Mix A+1µL of oligo 47 +2µL of DNA | ||
| + | * Mix B : | ||
| + | ** Mix A : 1.125mL | ||
| + | ** Oligo 43 : 25µL | ||
| + | *** NirB with RBS-LacZ-Term in pSB1C3 : | ||
| + | **** 23µL of Mix B+2µL of DNA | ||
| + | *** NirB with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 23µL of Mix B+2µL of DNA | ||
| + | *** NarG with RBS-LacZ-Term in pSB1C3 : | ||
| + | **** 23µL of Mix B+2µL of DNA | ||
| + | *** NarG with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 23µL of Mix B+2µL of DNA | ||
| - | + | *** NarK with RBS-LacZ-Term in pSB1C3 : | |
| + | **** 23µL of Mix B+2µL of DNA | ||
| + | *** NarK with RBS-Amil CP-Term in pSB1C3 : | ||
| + | **** 23µL of Mix B+2µL of DNA | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | PCR Program for NirB with RBS-LacZ-Term in pSB1C3, NarG with RBS-LacZ-Term in pSB1C3, NarK with RBS-LacZ-Term in pSB1C3 : | |
| + | |||
| + | [[File:PsPCRLacZ2108.jpg|400px]] | ||
| + | |||
| + | PCR Program for NirB with RBS-Amil CP-Term in pSB1C3, NarG with RBS-Amil CP-Term in pSB1C3, NarK with RBS-Amil CP-Term in pSB1C3 : | ||
| + | |||
| + | [[File:PsPCRAmilCP2108.jpg|400px]] | ||
| + | |||
| + | ====2 - Culture of BBa_K1155000==== | ||
| - | |||
Nguyen | Nguyen | ||
| - | + | We made culture of BBa_K115500 strains in 5ml LB with Chlorenphenicol. | |
| - | + | We incubate it at 37°C over night with shake 180 rpm. | |
| - | + | ===='''Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3'''==== | |
| - | + | ====1 - Electrophoresis of the digestion of BBa_J04450 by EcoRI/PstI==== | |
| - | + | ||
| - | ====1 - Electrophoresis of the digestion of | + | |
XiaoJing | XiaoJing | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel2108.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 17µL of | + | * Well 2 : 17µL of BBa_J04450 digested by EcoRI/PstI+3µL of H2O+4µL of 6X loading buffer |
* Gel : 1% | * Gel : 1% | ||
|} | |} | ||
| Line 74: | Line 94: | ||
Expected sizes : | Expected sizes : | ||
* PSB3K3 : 2750 bp | * PSB3K3 : 2750 bp | ||
| - | |||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained a fragment with the right size. We can purify it. |
|} | |} | ||
| + | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ||
| - | === | + | ====Objective : obtaining FNR and BphR2 proteins==== |
| - | == | + | |
| - | + | ||
| - | + | ====1 - Transformation of RBS-BphR2, FNR, RBS-FNR in DH5α==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | XiaoJing | ||
| + | Protocol : [[Team:Paris_Saclay/Protocols/Transformation|Bacterial transformation]] | ||
| + | {| border="1" align="center" | ||
| + | |[[Team:Paris Saclay/Notebook/August/20|<big>Previous day</big>]] | ||
| + | |[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | ||
| + | |[[Team:Paris Saclay/Notebook/August/22|<big>Next day</big>]] | ||
| + | |} | ||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Latest revision as of 01:16, 5 October 2013
Notebook : August 21
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
1 - Colony PCR of NarK, NarG or NirB with RBS-LacZ-Term or RBS-AmilCP-Term in pSB1C3 in DH5α
Damir, Xiaojing
|
The transformation was good. We will do a Colony PCR. |
Colonies repiquée dans 10µL d'eau. We do 8 PCR for each ligation.
Used quantities :
- Mix A :
- Buffer Dream Taq : 250µL
- dNTP : 50µL
- Oligo 44 : 50µL
- Dream Taq : 20µL
- H2O : 1.88mL
- NirB with RBS-LacZ-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 45+2µL of DNA
- NirB with RBS-Amil CP-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 45 +2µL of DNA
- NirB with RBS-LacZ-Term in pSB1C3 :
- NarG with RBS-LacZ-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 41+2µL of DNA
- NarG with RBS-Amil CP-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 41 +2µL of DNA
- NarG with RBS-LacZ-Term in pSB1C3 :
- NarK with RBS-LacZ-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 47+2µL of DNA
- NarK with RBS-Amil CP-Term in pSB1C3 :
- 45µL of Mix A+1µL of oligo 47 +2µL of DNA
- NarK with RBS-LacZ-Term in pSB1C3 :
- Mix B :
- Mix A : 1.125mL
- Oligo 43 : 25µL
- NirB with RBS-LacZ-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NirB with RBS-Amil CP-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NirB with RBS-LacZ-Term in pSB1C3 :
- NarG with RBS-LacZ-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NarG with RBS-Amil CP-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NarG with RBS-LacZ-Term in pSB1C3 :
- NarK with RBS-LacZ-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NarK with RBS-Amil CP-Term in pSB1C3 :
- 23µL of Mix B+2µL of DNA
- NarK with RBS-LacZ-Term in pSB1C3 :
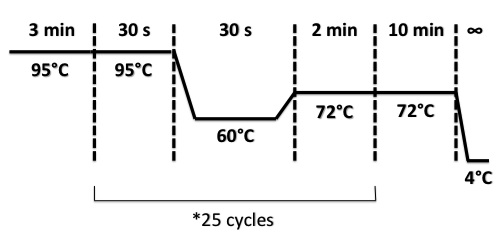
PCR Program for NirB with RBS-LacZ-Term in pSB1C3, NarG with RBS-LacZ-Term in pSB1C3, NarK with RBS-LacZ-Term in pSB1C3 :
PCR Program for NirB with RBS-Amil CP-Term in pSB1C3, NarG with RBS-Amil CP-Term in pSB1C3, NarK with RBS-Amil CP-Term in pSB1C3 :
2 - Culture of BBa_K1155000
Nguyen
We made culture of BBa_K115500 strains in 5ml LB with Chlorenphenicol. We incubate it at 37°C over night with shake 180 rpm.
Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3
1 - Electrophoresis of the digestion of BBa_J04450 by EcoRI/PstI
XiaoJing
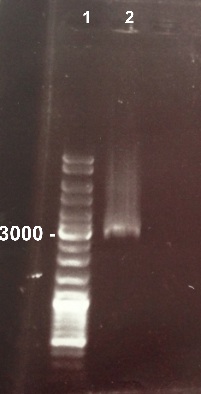
|
|
Expected sizes :
- PSB3K3 : 2750 bp
|
We obtained a fragment with the right size. We can purify it. |
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR and BphR2 proteins
1 - Transformation of RBS-BphR2, FNR, RBS-FNR in DH5α
XiaoJing
Protocol : Bacterial transformation
| Previous day | Back to calendar | Next day |
 "
"