Team:NYMU-Taipei/Project/Enter
From 2013.igem.org
| (45 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| + | =Entering of ''Bee. coli''= | ||
==Introduction== | ==Introduction== | ||
| - | In this part, the main purpose is to send ''Bee. coli'' into bees and make sure it can survive in the midguts. | + | In this part, the main purpose is to send ''Bee. coli'' into bees and make sure it can survive in the midguts. We chose ''Escherichia coli'' MG1655 as the chassis of our circuit, then incorporating MG1655 into alginate microcapsules, and fed bees with sugar water containing microencapsulation beads. According to our experiment, this method could successfully transport the engineered ''Bee. coli'' into the bees’ midgut. |
==Background== | ==Background== | ||
| - | ===Why did we choose MG1655=== | + | <!-- |
| + | ===Why did we choose to use MG1655=== | ||
[[File:NYMU_Bacteria_exist_in_honeybees'_midguts_naturally.png|thumb|300px|right|'''Bacteria exist in honeybees' midguts naturally'''<br>Babendreier, D., Joller, D., Romeis, J., Bigler, F., & Widmer, F. (March 01, 2007). Bacterial community structures in honeybee 48 | [[File:NYMU_Bacteria_exist_in_honeybees'_midguts_naturally.png|thumb|300px|right|'''Bacteria exist in honeybees' midguts naturally'''<br>Babendreier, D., Joller, D., Romeis, J., Bigler, F., & Widmer, F. (March 01, 2007). Bacterial community structures in honeybee 48 | ||
intestines and their response to two insecticidal proteins. Fems Microbiology Ecology, 59, 3, 600-610]] | intestines and their response to two insecticidal proteins. Fems Microbiology Ecology, 59, 3, 600-610]] | ||
| - | #The main reason we want to choose one of the substrain, MG1655, of ''E. coli'' | + | #The main reason we want to choose one of the substrain, MG1655, of ''E. coli'' K-12 strain as our carrier is ''E. coli'' K-12 strain exists in the midgut's of honeybees naturally. In addition, compared to nother substrain of K-12, MG1655 was selected for having few genetic manipulations from the archetypal E. coli K-12 strain, and it has already been well-studied as well. |
| - | #The second reason for choosing ''E. coli'' | + | #The second reason for choosing ''E. coli'' K-12 strain is that what we utilize to repress the sprouting of ''Nosema'' (Mannosidase), sense the invasion of ''Nosema'' (ROS) and kill ''Nosema'' (Defensin and Abaecin) are harmful to the microbes. Notably, they are naturally produced in bees as well. Besides, the immune system of honeybee will prevent our ''E. kobee'' colonizing in the epithelial cells of guts. To solve these problems, we select MG1655 which naturally exists in bees. |
#Therefore, MG1655 is supposed to survive in the midguts after we send it into bees by encapsulation. | #Therefore, MG1655 is supposed to survive in the midguts after we send it into bees by encapsulation. | ||
| - | #However, the transformation efficiency of MG1655 is not as good as that of DH5 alpha, which is another substrain of | + | #However, the transformation efficiency of MG1655 is not as good as that of DH5 alpha, which is another substrain of K-12. On the other hand, the oxidative stress tolerance of MG1655 is much better than DH5 alpha theoretically because it's more similar to wild-type. |
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| + | --> | ||
| - | |||
| - | |||
| - | |||
| - | + | ===Why encapsulation?=== | |
| + | The host immune system imposes a great threat on our engineered ''E.coli''. Besides we have to transported our engineered ''E.coli'' in the bees through bees’ digest system, which may contain many kinds of digestive solution with variable pH value that do harm to our engineered ''E.coli''. | ||
| - | + | To solve this problem, we have to cover the ''E.coli'' with a membrane. And encapsulation is an easy way to reach the goal. Comparing to other approaches, cell encapsulation is easy to manipulate. So we choose cell encapsulation as the approach to cover the ''E.coli''. | |
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| - | === | + | ===Materials=== |
| - | + | There are many different biomaterials for cell encapsulation. Among those biomaterials, we chose alginate as our encapsulation material for its abundance, excellent biocompatibility and biodegradability properties. | |
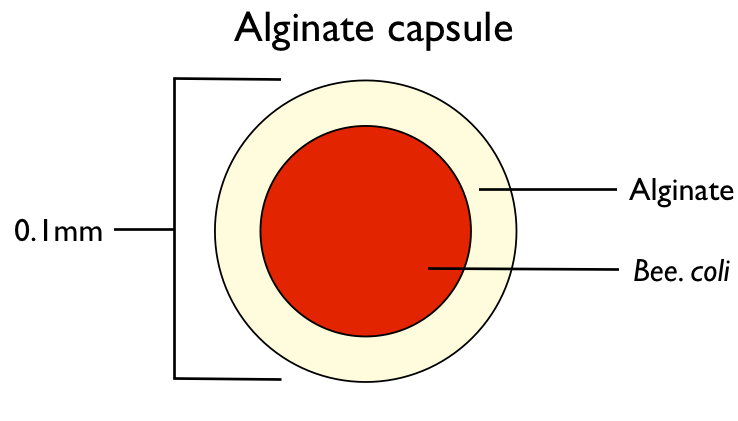
| - | + | We successfully encapsulated the engineered ''E.coli''( ''E.coli'' with GFP gene), and controlled its size under 0.1mm. Then we tried to transfer the engineered ''E.coli'' into bees midgut by adding some beads into their food. | |
| - | We | + | |
| + | {| | ||
| + | |- | ||
| + | ! Alginate structure | ||
| + | ! The structure of alginate capsule | ||
| + | |- | ||
| + | | [[File:NYMU_alginate1.png.png|thumb|right|300px]] | ||
| + | | [[File:NYMU_alginate_structure1.png|thumb|right|300px]] | ||
| + | |} | ||
| + | |||
| + | ===CCD-curing pellet=== | ||
| + | |||
| + | After we proved that microencapsulation of ''E.coli'' could be sent into midgut through bees’ digestion system, we came up with an idea that we can create a drug-like product which is the mixture of sugar and microencapsulation of engineered ''E.coli''. Therefore, we mixed the solution of microencapsulation with sugar cube and dry it. Then, we create the drug pellet which can cure CCD. | ||
| + | <div style="clear:both;"></div> | ||
| - | == | + | ==Experiments== |
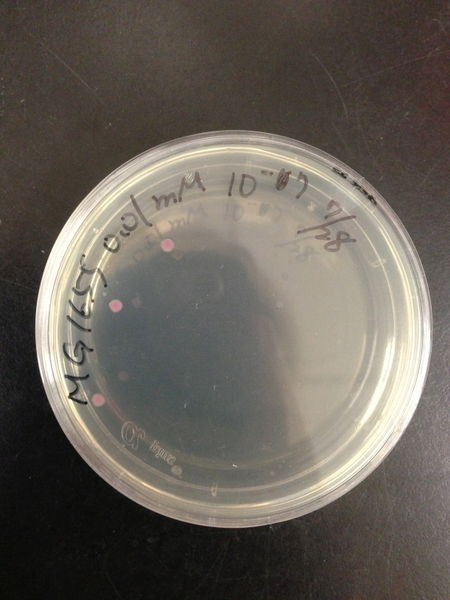
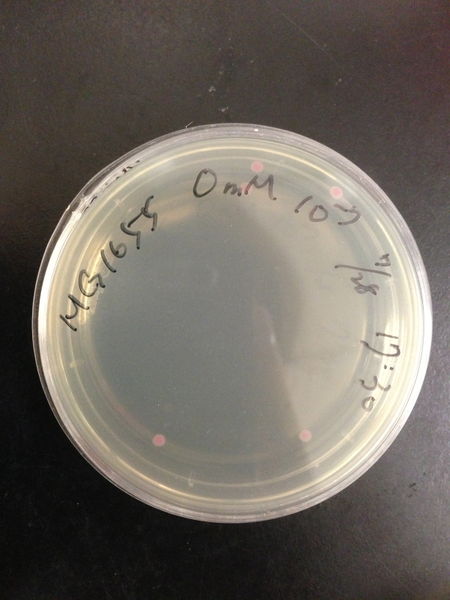
===Comparison of DH5 alpha and MG1655=== | ===Comparison of DH5 alpha and MG1655=== | ||
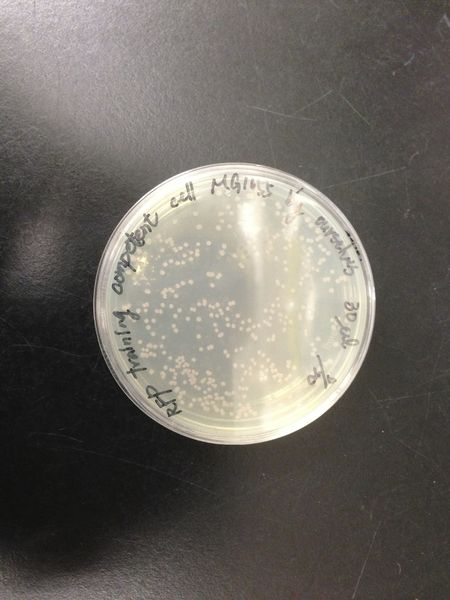
| - | We | + | We transformed red fluorescent protein into MG1655 and DH5 alpha, and then spread these bacteria onto the LB agar plates. After cultured at 37 degree for 16 hours, we calculated the number of colonies and found the transformation efficiency of DH5 alpha is about 100-fold higher than MG1655. |
{| | {| | ||
|- | |- | ||
| Line 47: | Line 61: | ||
===Oxidative stress tolerance of MG1655=== | ===Oxidative stress tolerance of MG1655=== | ||
| - | To know the oxidative stress tolerance of MG1655, we | + | To know the oxidative stress tolerance of MG1655, we tested the susceptibility to H<sub>2</sub>O<sub>2</sub> in MG1655.<br> |
| - | + | ====Survival test==== | |
#MG1655 cells were cultivated at 37 ˚C until the optical density (OD600) of the medium became 0.4~0.6 | #MG1655 cells were cultivated at 37 ˚C until the optical density (OD600) of the medium became 0.4~0.6 | ||
| - | #The culture was equally dispensed (1 ml each) into sample tubes, and then cells were treated with various concentrations of | + | #The culture was equally dispensed (1 ml each) into sample tubes, and then cells were treated with various concentrations of H<sub>2</sub>O<sub>2</sub> (final concentrations were as follows: 10μM, 50μM, 100μM, 500μM, 1 mM, 5mM, 10mM, 50mM and 100mM) at 37 ˚C for 60 min |
#Testing the OD of each samples | #Testing the OD of each samples | ||
#Doing serial dilution on samples by mixing fresh LB medium with samples | #Doing serial dilution on samples by mixing fresh LB medium with samples | ||
#20μl of the diluted sample was spread out on a LB plate (amp-) and incubated at 37 ˚C (overnight) <br> | #20μl of the diluted sample was spread out on a LB plate (amp-) and incubated at 37 ˚C (overnight) <br> | ||
| - | + | ====Results and discussion==== | |
{| | {| | ||
|- | |- | ||
| Line 65: | Line 79: | ||
|} | |} | ||
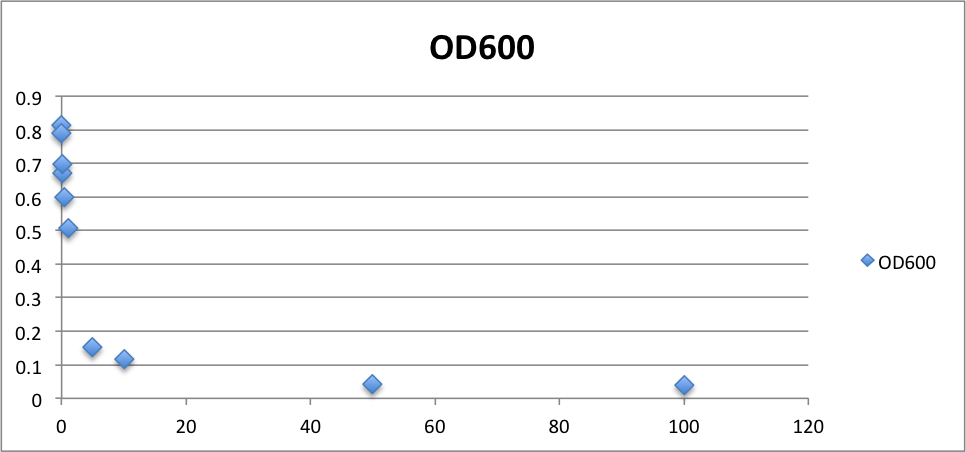
| - | From these charts, | + | From these charts, the oxidative stress tolerance of MG1655 is higher than 5mM, and epithelial cells is only 0.05mM. In the other words, the tolerance to ROS of MG1655 is over 100-fold higher than cells in midgut. Therefore, MG1655 will survive even in the stress environment in the midgut. |
{| | {| | ||
| Line 123: | Line 137: | ||
| - | === | + | ===Entering the bees' midgut=== |
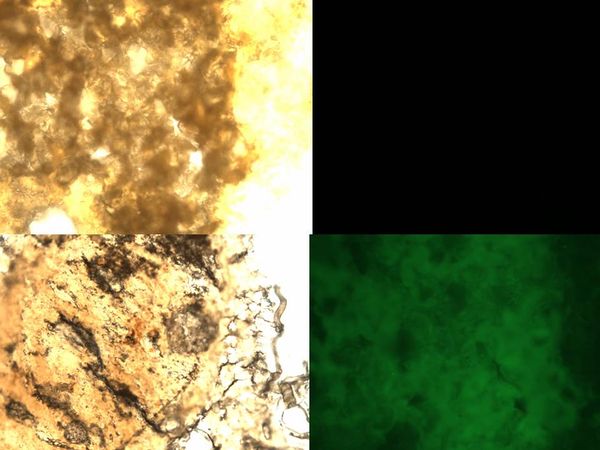
| - | [[Image: | + | ====Getting into bees’ midgut==== |
| - | We | + | [[Image:NYMU_TAIPEI_ENTERING!!.jpg|thumb|600px|right|left up: bees' gut , right up: no fluorescent light, left down: bees' gut after intaking ''Bee. coli'', right down: lots of fluorescent signal]] |
| + | We dropped some solution with microencapsulation of ''E.coli'' on the sugar cube. After three days, we pulled out bees’ midgut and see if there was engineered ''E.coli''. | ||
| + | We also pulled out bees’ midgut from those who did not intake engineered ''E.coli'' and compared the result. | ||
| + | <div style="clear:both;"></div> | ||
| - | + | ====Products==== | |
| + | [[Image:NYMU_TAIPEI_productentering.jpg|thumb|600px|right|left up: bees' gut , right up: no fluorescent light, left down: bees' gut after intaking our product, right down: lots of fluorescent signal]] | ||
| + | After the engineered ''E.coli'' was made into our pellet product, we wondered that if the engineered ''E.coli'' still work. | ||
| + | We dissolved our pellet product in sugar water and fed our bees. After 3 days, we pulled out bees’ midgut and see if there was any engineered ''E.coli''. The result show that there were still engineered ''E.coli'', that was our product really work. | ||
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| - | [[Image: | + | ==Outcome== |
| - | + | [[Image:NYMU_TAIPEI_product.jpg|right|The sugar cube with engineered ''E.coli'' in it. ]] | |
| - | + | We selected MG1655 as the chassis of our circuit, then successfully finding a way which can avoid the threat of immune system and ensure that ''Bee. coli'' won't spread into the environment while transporting ''Bee. coli'' into bees. Besides, once our chassis entered the midguts of bees, it is able to survive and fight against ''Nosema''. | |
| + | We mixed the solution of microencapsulation beads with sugar pellet and dried it. That's what we called CCD-curing pellet. | ||
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| - | + | {{:Team:NYMU-Taipei/Footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | {{: | + | |
Latest revision as of 17:46, 27 October 2013


Contents |
Entering of Bee. coli
Introduction
In this part, the main purpose is to send Bee. coli into bees and make sure it can survive in the midguts. We chose Escherichia coli MG1655 as the chassis of our circuit, then incorporating MG1655 into alginate microcapsules, and fed bees with sugar water containing microencapsulation beads. According to our experiment, this method could successfully transport the engineered Bee. coli into the bees’ midgut.
Background
Why encapsulation?
The host immune system imposes a great threat on our engineered E.coli. Besides we have to transported our engineered E.coli in the bees through bees’ digest system, which may contain many kinds of digestive solution with variable pH value that do harm to our engineered E.coli.
To solve this problem, we have to cover the E.coli with a membrane. And encapsulation is an easy way to reach the goal. Comparing to other approaches, cell encapsulation is easy to manipulate. So we choose cell encapsulation as the approach to cover the E.coli.
Materials
There are many different biomaterials for cell encapsulation. Among those biomaterials, we chose alginate as our encapsulation material for its abundance, excellent biocompatibility and biodegradability properties.
We successfully encapsulated the engineered E.coli( E.coli with GFP gene), and controlled its size under 0.1mm. Then we tried to transfer the engineered E.coli into bees midgut by adding some beads into their food.
| Alginate structure | The structure of alginate capsule |
|---|---|
CCD-curing pellet
After we proved that microencapsulation of E.coli could be sent into midgut through bees’ digestion system, we came up with an idea that we can create a drug-like product which is the mixture of sugar and microencapsulation of engineered E.coli. Therefore, we mixed the solution of microencapsulation with sugar cube and dry it. Then, we create the drug pellet which can cure CCD.
Experiments
Comparison of DH5 alpha and MG1655
We transformed red fluorescent protein into MG1655 and DH5 alpha, and then spread these bacteria onto the LB agar plates. After cultured at 37 degree for 16 hours, we calculated the number of colonies and found the transformation efficiency of DH5 alpha is about 100-fold higher than MG1655.
| DH5 alpha | MG1655 |
|---|---|
Oxidative stress tolerance of MG1655
To know the oxidative stress tolerance of MG1655, we tested the susceptibility to H2O2 in MG1655.
Survival test
- MG1655 cells were cultivated at 37 ˚C until the optical density (OD600) of the medium became 0.4~0.6
- The culture was equally dispensed (1 ml each) into sample tubes, and then cells were treated with various concentrations of H2O2 (final concentrations were as follows: 10μM, 50μM, 100μM, 500μM, 1 mM, 5mM, 10mM, 50mM and 100mM) at 37 ˚C for 60 min
- Testing the OD of each samples
- Doing serial dilution on samples by mixing fresh LB medium with samples
- 20μl of the diluted sample was spread out on a LB plate (amp-) and incubated at 37 ˚C (overnight)
Results and discussion
| Susceptibility to H2O2 in MG1655 shown in OD | Susceptibility to H2O2 in MG1655 shown in colonies |
|---|---|
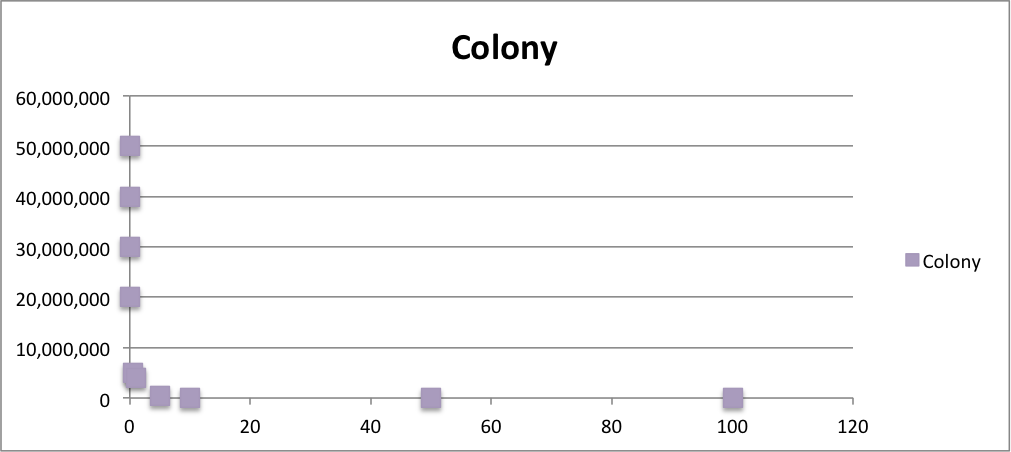
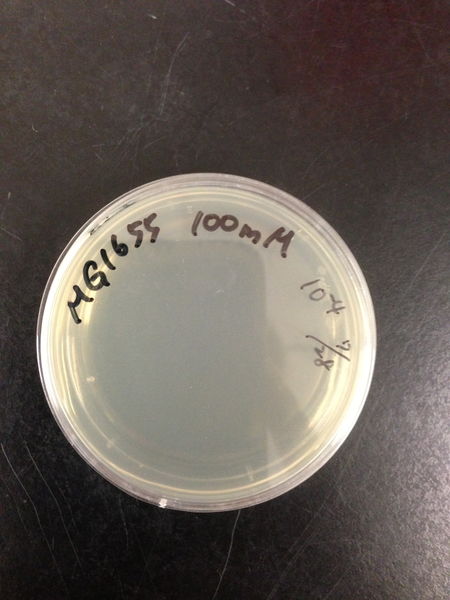
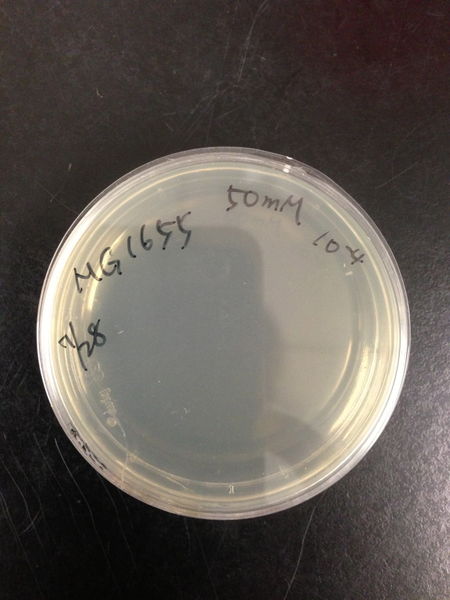
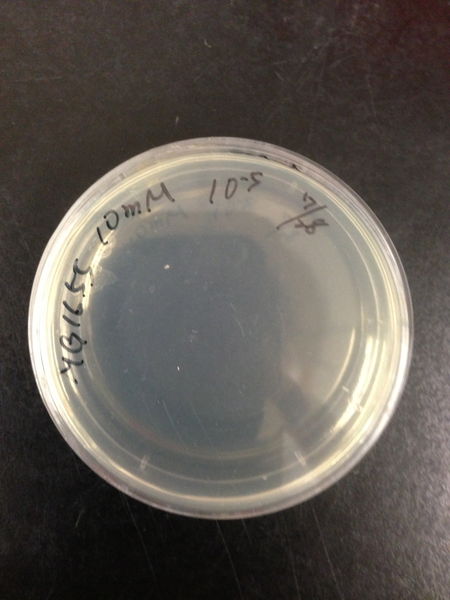
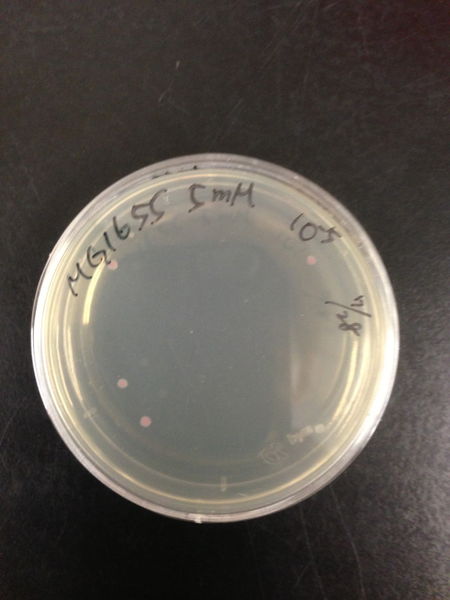
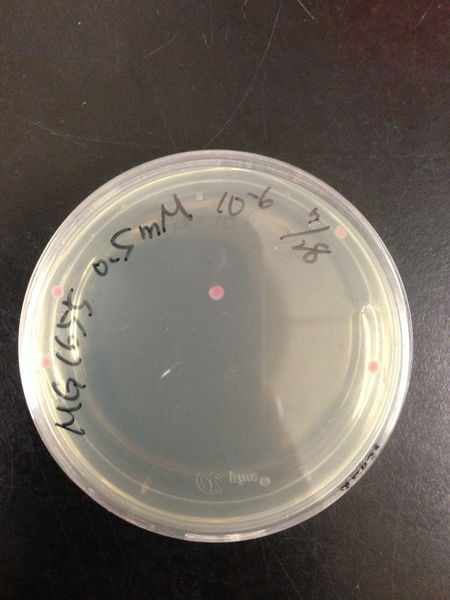
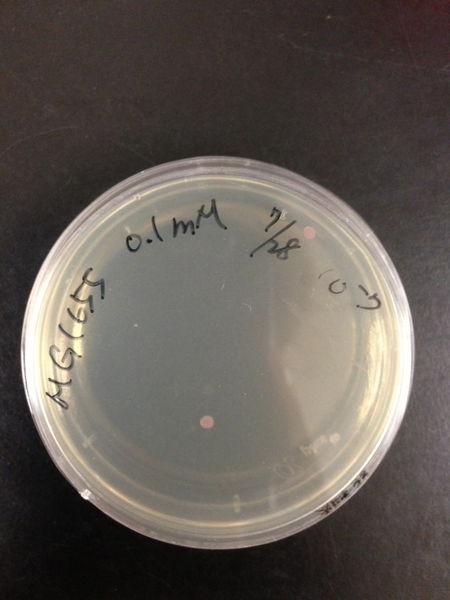
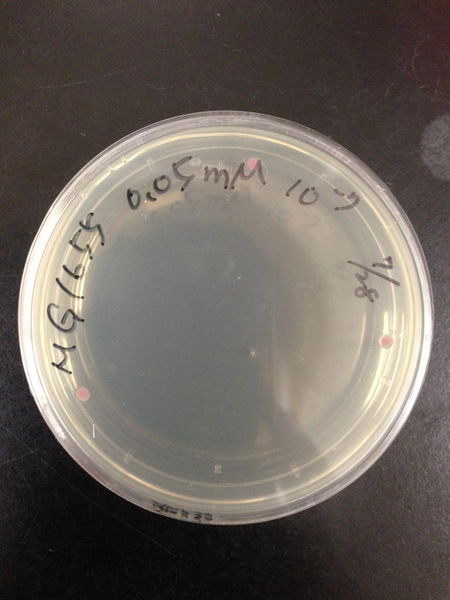
From these charts, the oxidative stress tolerance of MG1655 is higher than 5mM, and epithelial cells is only 0.05mM. In the other words, the tolerance to ROS of MG1655 is over 100-fold higher than cells in midgut. Therefore, MG1655 will survive even in the stress environment in the midgut.
| Treated with 100mM H2O2 diluted 104times | Treated with 50mM H2O2 diluted 104times | Treated with 10mM H2O2 diluted 105times | Treated with 5mM H2O2 diluted 105times | Treated with 1mM H2O2 diluted 106times |
|---|---|---|---|---|
| Treated with 500μM H2O2 diluted 106times | Treated with 100μM H2O2 diluted 107times | Treated with 50μM H2O2 diluted 107times | Treated with 10μM H2O2 diluted 107times | Treated with 0μM H2O2 diluted 107times |
Comparison table of MG1655 and DH5 alpha
| Comparison | MG1655 | DH5 alpha |
|---|---|---|
| Strain | K12 | K12 |
| Genotype | F-, lambda-, rph-1 | -fhuA2 lac(del)U169 phoA glnV44 Φ80' lacZ(del)M15 gyrA96 recA1 relA1 endA1 |
| Characteristics | Similar to wild-type | Designed for transformation |
| Transformation efficiency | >1x108cfu/μg | >1x106cfu/μg |
| Oxidative stress tolerance | <0.1mM | >5mM |
Entering the bees' midgut
Getting into bees’ midgut
We dropped some solution with microencapsulation of E.coli on the sugar cube. After three days, we pulled out bees’ midgut and see if there was engineered E.coli. We also pulled out bees’ midgut from those who did not intake engineered E.coli and compared the result.
Products
After the engineered E.coli was made into our pellet product, we wondered that if the engineered E.coli still work. We dissolved our pellet product in sugar water and fed our bees. After 3 days, we pulled out bees’ midgut and see if there was any engineered E.coli. The result show that there were still engineered E.coli, that was our product really work.
Outcome
We selected MG1655 as the chassis of our circuit, then successfully finding a way which can avoid the threat of immune system and ensure that Bee. coli won't spread into the environment while transporting Bee. coli into bees. Besides, once our chassis entered the midguts of bees, it is able to survive and fight against Nosema. We mixed the solution of microencapsulation beads with sugar pellet and dried it. That's what we called CCD-curing pellet.
 "
"