Team:NYMU-Taipei/Project/Inhibition/Killing
From 2013.igem.org
Gastbyqpzm (Talk | contribs) |
|||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| + | =Killing ''Nosema ceranae''= | ||
==Introduction== | ==Introduction== | ||
| - | <p> | + | <p>Our main goal is to kill ''Nosema ceranae'' without harming the bees.</p> |
| - | + | <p>First, we came up with several chemicals that might achieve our goal. They are antimicrobial peptides, such as Apidaecin, [http://parts.igem.org/Part:BBa_K1104300 Abaecin], Hymenoptaecin, [http://parts.igem.org/Part:BBa_K1104301 Defensin1], which are all antimicrobial peptides in bees. Because these chemicals are secreted by bees themselves, they definitely do no harm to bees. Among four kinds of antimicrobial peptides, Abaecin and Defensin are much smaller in terms of their molecular sizes so it would be easier for us to biosynthesize them, that was the first reason why we chose [http://parts.igem.org/Part:BBa_K1104300 Abaecin] and [http://parts.igem.org/Part:BBa_K1104301 Defensin1] to treat bees. The second reason was that the sequence of Apidaecin cannot be found at this moment. The third reason is that [http://parts.igem.org/Part:BBa_K1104300 Abaecin] and [http://parts.igem.org/Part:BBa_K1104301 Defensin1] have been relatively well-studied. In addition to antimicrobial peptides, we figured out other chemicals, which are Chitinase, Herein, Protofil, Cytochrome P450. Unfortunately, Chitinase, Herein, Protofil, and Cytochrome P450 have several defects. For example, they are all macromolecules, which make them difficult to be synthesized by ''E. coli''. Moreover, even if ''E. coli'' can successfully synthesize them, it is still too hard to make these chemicals be released out of the cells. Based on all of the above reasons, we finally decided to use [http://parts.igem.org/Part:BBa_K1104301 Defensin1] and [http://parts.igem.org/Part:BBa_K1104300 Abaecin] to treat the bees infected by ''Nosema ceranae''.</p> | |
| - | <p> | + | |
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| Line 11: | Line 11: | ||
==='''How Do AntiMicrobial Peptides Work?'''=== | ==='''How Do AntiMicrobial Peptides Work?'''=== | ||
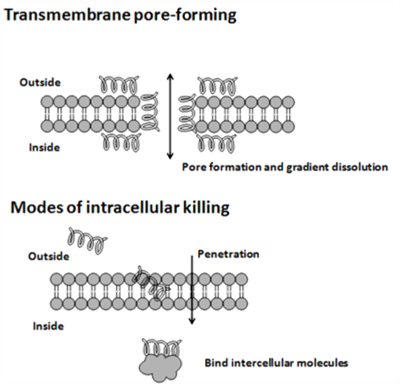
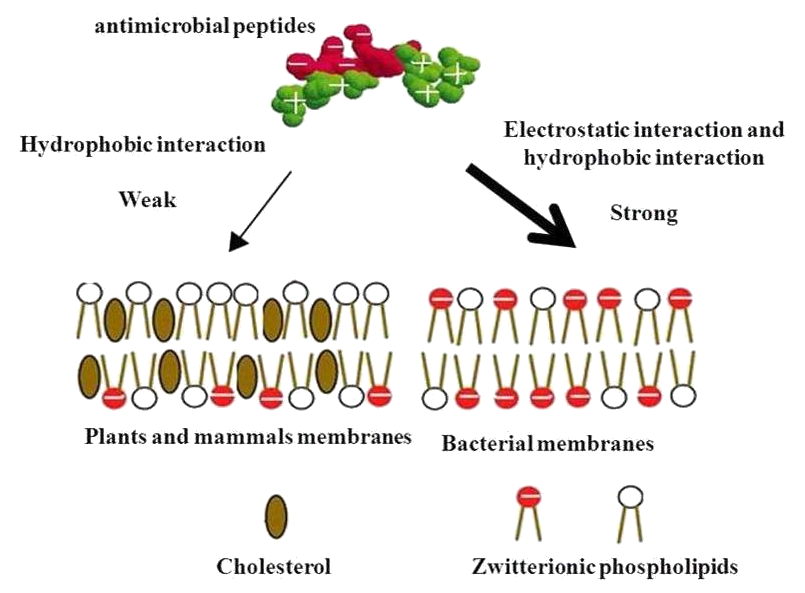
| - | <p> | + | <p>The cytoplasmic membrane is a frequent target, but peptides may also interfere with DNA and protein synthesis, protein folding, and cell wall synthesis. The initial contact between the peptide and the target organism is electrostatic, as most bacterial surfaces are anionic, or hydrophobic. Their amino acid composition, amphipathicity, cationic charge and size allow them to attach to and insert into membrane bilayers to form pores by ‘barrel-stave’, ‘carpet’ or ‘toroidal-pore’ mechanisms. Alternately, they may penetrate into the cell to bind intracellular molecules which are crucial to cell living. Intracellular binding models includes inhibition of cell wall synthesis, alteration of the cytoplasmic membrane, activation of autolysin, inhibition of DNA, RNA, and protein synthesis, and inhibition of certain enzymes. One emerging technique for the study of such mechanisms is dual polarisation interferometry. In contrast to many conventional antibiotics these peptides appear to be bactericidal instead of bacteriostatic.</p> |
[[File:NYMU_Group6_AMP-1.png|350px]] [[File:NYMU Group6 AMP-2.png|350px]] | [[File:NYMU_Group6_AMP-1.png|350px]] [[File:NYMU Group6 AMP-2.png|350px]] | ||
| Line 17: | Line 17: | ||
*See more("articles & pictures" modified from) [http://en.wikipedia.org/wiki/Antimicrobial_peptides http://en.wikipedia.org/wiki/Antimicrobial_peptides] | *See more("articles & pictures" modified from) [http://en.wikipedia.org/wiki/Antimicrobial_peptides http://en.wikipedia.org/wiki/Antimicrobial_peptides] | ||
| - | <p> | + | <p>After deciding the chemicals that are able to kill Nosema ceranae without harming the bees and how they work, we designed our experiment. The following is our material list as well as experimental procedure.</p> |
=='''Circuit Design & Experimental Methods'''== | =='''Circuit Design & Experimental Methods'''== | ||
| Line 27: | Line 27: | ||
===Experimental Methods=== | ===Experimental Methods=== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | |||
| + | '''Testing the influence of antimicrobial peptides on ''Bee. coli''''' | ||
| + | <p>1. Make experiment group and control group with our ''Bee. coli''</p> | ||
| + | <p> (Experiment''':'''transformed with the Defensin1 gene circuit;Control''':''' w/o the gene)</p> | ||
| + | <p>2. Determine both groups' OD 600 for three to five days</p> | ||
| + | <p>3. Collect and Analyze the data and make growth-rate graphs to check whether Defensin1 would kill or inhibit</p> | ||
| + | <p> ''Bee. coli''</p> | ||
| + | <p>4. Change the gene circuit into the Abaecin one and do steps 1~3 in parallel with the Defensin1 experiments</p> | ||
| + | |||
| + | |||
| + | '''Check the efficiency of our products to against Nosema''' | ||
| + | |||
| + | <p>1. Make experiment group and control group with our ''Bee. coli''</p> | ||
| + | <p> (Experiment''':'''transformed with the Defensin1 gene circuit;Control''':''' w/o the gene)</p> | ||
| + | <p>2. Both add 5x10³ spores in the liquid for experiment and control groups, respectively</p> | ||
| + | <p>3. Around three days after step 2, collect data for three to six days</p> | ||
| + | <p> (Determine the change amount of ''Nosema'' spores through RBC counting chamber)</p> | ||
| + | <p>4. Analyze all the information and make growth-rate graphs to check Defensin1's killing ability</p> | ||
| + | <p>5. Change the gene circuit into the Abaecin one and do steps 1~3 in parallel with the Defensin1 experiments</p> | ||
| + | |||
| + | ==Defensin1 Function Test== | ||
| + | <p>[[File:NYMU Nosema DNA Gel.PNG|thumb|300px|Electrophoresis of the Supernatant PCR production]]</p> | ||
| + | In this case, we want to see whether our product - Defensin1 can function or not. Combined with some good work done from "Prohibiting Sprouting", we can prove its function through running the PCR to check whether there is any ''Nosema ceranae'' spores' DNA in the Supernatant rather than in the bodies of spores. That is to say, our defensin can really work to penetrate the cell membranes of the spores and let the DNA come out. | ||
| + | As the photo it shows, it is very clear that only spores treated with the MG1655-D lysate can show the band in electrophoresis, in spite of the not-so-perfect bands. | ||
| + | |||
| + | ==Defensin1 Resistance of MG1655 Test== | ||
| + | |||
| + | In this experiment, our main goal is to test the resistance to defensin1 of ''E. coli'' K12 MG1655. If MG1655 can have resistance against Defensin1, we can make sure that our ''Bee. coli'' wouldn’t kill itself. First we clone the defensin1 production gene into MG1655 (J23102+RBS+K1104301+Terminator), this engineered ''E. coli'' will be called MG1655-D for abbreviation latter. We use ultrasonication to break MG1655-D and MG1655 wildtype(as blank). Add these lysate into MG1655 liquid, culture at 37℃, observe if the growth of MG1655 will be influenced by MG1655-D lysate. | ||
| + | === Plating Test === | ||
| + | We choose 3 concentrations each lysate, 3 time point, do plating and count the colony amount to observe the growth of MG1655. | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! colony amount !! 50λ MG1655-D lysate !! 100λ MG1655-D lysate !! 150λ MG1655-D lysate !! 50λ MG1655 lysate !! 100λ MG1655 lysate !! 150λ MG1655 lysate | ||
| + | |- | ||
| + | | 60 mins || 716 || 1188 || 2092 || 1836 || 2608 || 5544 | ||
| + | |- | ||
| + | | 90 mins || 1284 || 1428 || 2044 || 2496 || 2940 || 5112 | ||
| + | |- | ||
| + | | 120 mins || 2076 || 1984 || 2388 || 2772 || 3840 || 7680 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | [[File:NYMU_Plating_test_result.PNG|500px]] | ||
| + | |||
| + | Since the colony amount is too much to count, we divide the plate into a quarter, estimate the colony amount. Because we need to include more concentration and keep track of the growth of MG1655 longer, also concerning the precision of counting colony amount on the agar plate, we decide to do the second round of this experiment, the 96-well plate OD600 test. | ||
| + | |||
| + | === 96-well plate OD600 Test === | ||
| + | We choose 4 concentrations each lysate, culture it in the 96-well plate, keep track of its OD600 every 30 minutes till 480 minutes. | ||
| + | |||
| + | |||
| + | <p>[[File:NYMU MG1655 Def1 resistance.PNG|800px]]</p> | ||
| + | <p>[[File:NYMU 4-in-1.PNG|800px]]</p> | ||
| + | <p>[[File:NYMU Nutrient equal.PNG|800px]]</p> | ||
| + | |||
| + | In conclusion, it seems that the MG1655-D lysate doesn’t have, or have a little effect on the growth of MG1655 in LB liquid culture. In the 96-well plate OD600 test, MG1655-D lysate just depress the growth of MG1655 a little compare to MG1655 lysate. | ||
| + | That is, our ''Bee. coli'' wouldn’t harm itself, not to mention suicide. | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 21:19, 28 October 2013


Contents |
Killing Nosema ceranae
Introduction
Our main goal is to kill Nosema ceranae without harming the bees.
First, we came up with several chemicals that might achieve our goal. They are antimicrobial peptides, such as Apidaecin, [http://parts.igem.org/Part:BBa_K1104300 Abaecin], Hymenoptaecin, [http://parts.igem.org/Part:BBa_K1104301 Defensin1], which are all antimicrobial peptides in bees. Because these chemicals are secreted by bees themselves, they definitely do no harm to bees. Among four kinds of antimicrobial peptides, Abaecin and Defensin are much smaller in terms of their molecular sizes so it would be easier for us to biosynthesize them, that was the first reason why we chose [http://parts.igem.org/Part:BBa_K1104300 Abaecin] and [http://parts.igem.org/Part:BBa_K1104301 Defensin1] to treat bees. The second reason was that the sequence of Apidaecin cannot be found at this moment. The third reason is that [http://parts.igem.org/Part:BBa_K1104300 Abaecin] and [http://parts.igem.org/Part:BBa_K1104301 Defensin1] have been relatively well-studied. In addition to antimicrobial peptides, we figured out other chemicals, which are Chitinase, Herein, Protofil, Cytochrome P450. Unfortunately, Chitinase, Herein, Protofil, and Cytochrome P450 have several defects. For example, they are all macromolecules, which make them difficult to be synthesized by E. coli. Moreover, even if E. coli can successfully synthesize them, it is still too hard to make these chemicals be released out of the cells. Based on all of the above reasons, we finally decided to use [http://parts.igem.org/Part:BBa_K1104301 Defensin1] and [http://parts.igem.org/Part:BBa_K1104300 Abaecin] to treat the bees infected by Nosema ceranae.
Background
How Do AntiMicrobial Peptides Work?
The cytoplasmic membrane is a frequent target, but peptides may also interfere with DNA and protein synthesis, protein folding, and cell wall synthesis. The initial contact between the peptide and the target organism is electrostatic, as most bacterial surfaces are anionic, or hydrophobic. Their amino acid composition, amphipathicity, cationic charge and size allow them to attach to and insert into membrane bilayers to form pores by ‘barrel-stave’, ‘carpet’ or ‘toroidal-pore’ mechanisms. Alternately, they may penetrate into the cell to bind intracellular molecules which are crucial to cell living. Intracellular binding models includes inhibition of cell wall synthesis, alteration of the cytoplasmic membrane, activation of autolysin, inhibition of DNA, RNA, and protein synthesis, and inhibition of certain enzymes. One emerging technique for the study of such mechanisms is dual polarisation interferometry. In contrast to many conventional antibiotics these peptides appear to be bactericidal instead of bacteriostatic.
- See more("articles & pictures" modified from) [http://en.wikipedia.org/wiki/Antimicrobial_peptides http://en.wikipedia.org/wiki/Antimicrobial_peptides]
After deciding the chemicals that are able to kill Nosema ceranae without harming the bees and how they work, we designed our experiment. The following is our material list as well as experimental procedure.
Circuit Design & Experimental Methods
Circuit Design
[http://parts.igem.org/Part:BBa_K1104300 Abaecin]
[http://parts.igem.org/Part:BBa_K1104301 Defensin1]
Experimental Methods
Testing the influence of antimicrobial peptides on Bee. coli
1. Make experiment group and control group with our Bee. coli
(Experiment:transformed with the Defensin1 gene circuit;Control: w/o the gene)
2. Determine both groups' OD 600 for three to five days
3. Collect and Analyze the data and make growth-rate graphs to check whether Defensin1 would kill or inhibit
Bee. coli
4. Change the gene circuit into the Abaecin one and do steps 1~3 in parallel with the Defensin1 experiments
Check the efficiency of our products to against Nosema
1. Make experiment group and control group with our Bee. coli
(Experiment:transformed with the Defensin1 gene circuit;Control: w/o the gene)
2. Both add 5x10³ spores in the liquid for experiment and control groups, respectively
3. Around three days after step 2, collect data for three to six days
(Determine the change amount of Nosema spores through RBC counting chamber)
4. Analyze all the information and make growth-rate graphs to check Defensin1's killing ability
5. Change the gene circuit into the Abaecin one and do steps 1~3 in parallel with the Defensin1 experiments
Defensin1 Function Test
In this case, we want to see whether our product - Defensin1 can function or not. Combined with some good work done from "Prohibiting Sprouting", we can prove its function through running the PCR to check whether there is any Nosema ceranae spores' DNA in the Supernatant rather than in the bodies of spores. That is to say, our defensin can really work to penetrate the cell membranes of the spores and let the DNA come out. As the photo it shows, it is very clear that only spores treated with the MG1655-D lysate can show the band in electrophoresis, in spite of the not-so-perfect bands.
Defensin1 Resistance of MG1655 Test
In this experiment, our main goal is to test the resistance to defensin1 of E. coli K12 MG1655. If MG1655 can have resistance against Defensin1, we can make sure that our Bee. coli wouldn’t kill itself. First we clone the defensin1 production gene into MG1655 (J23102+RBS+K1104301+Terminator), this engineered E. coli will be called MG1655-D for abbreviation latter. We use ultrasonication to break MG1655-D and MG1655 wildtype(as blank). Add these lysate into MG1655 liquid, culture at 37℃, observe if the growth of MG1655 will be influenced by MG1655-D lysate.
Plating Test
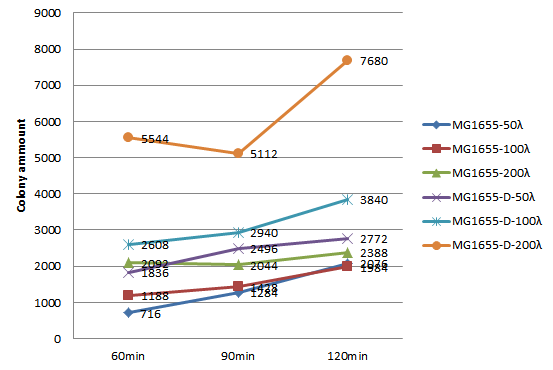
We choose 3 concentrations each lysate, 3 time point, do plating and count the colony amount to observe the growth of MG1655.
| colony amount | 50λ MG1655-D lysate | 100λ MG1655-D lysate | 150λ MG1655-D lysate | 50λ MG1655 lysate | 100λ MG1655 lysate | 150λ MG1655 lysate |
|---|---|---|---|---|---|---|
| 60 mins | 716 | 1188 | 2092 | 1836 | 2608 | 5544 |
| 90 mins | 1284 | 1428 | 2044 | 2496 | 2940 | 5112 |
| 120 mins | 2076 | 1984 | 2388 | 2772 | 3840 | 7680 |
Since the colony amount is too much to count, we divide the plate into a quarter, estimate the colony amount. Because we need to include more concentration and keep track of the growth of MG1655 longer, also concerning the precision of counting colony amount on the agar plate, we decide to do the second round of this experiment, the 96-well plate OD600 test.
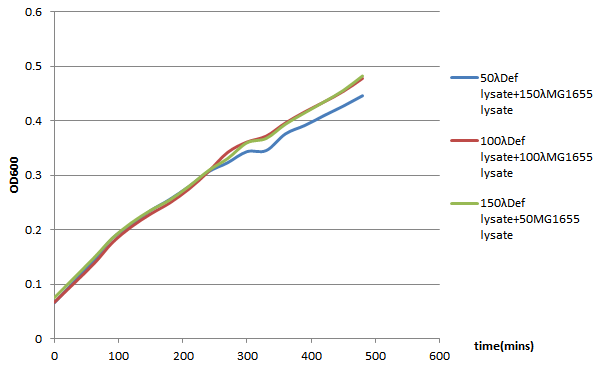
96-well plate OD600 Test
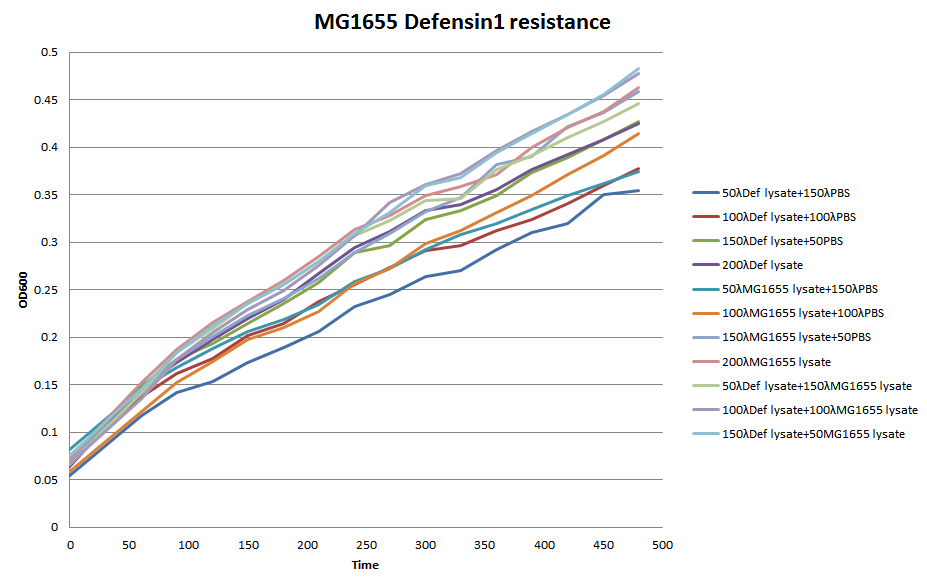
We choose 4 concentrations each lysate, culture it in the 96-well plate, keep track of its OD600 every 30 minutes till 480 minutes.
In conclusion, it seems that the MG1655-D lysate doesn’t have, or have a little effect on the growth of MG1655 in LB liquid culture. In the 96-well plate OD600 test, MG1655-D lysate just depress the growth of MG1655 a little compare to MG1655 lysate. That is, our Bee. coli wouldn’t harm itself, not to mention suicide.
 "
"