Team:NYMU-Taipei/Project/safe
From 2013.igem.org
| (21 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
===Blue Light Sensing Device=== | ===Blue Light Sensing Device=== | ||
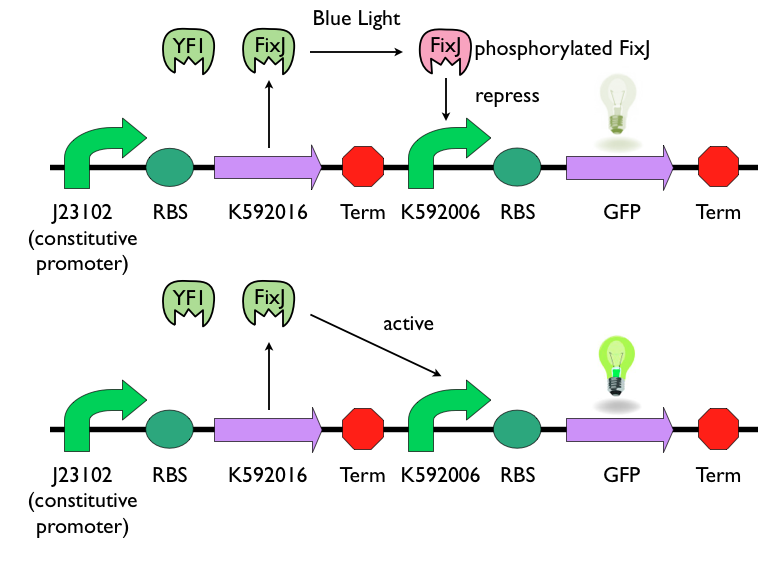
| - | We chose to use [http://parts.igem.org/Part:BBa_K592016 K592016] as our light sensing device. [http://parts.igem.org/Part:BBa_K592016 K592016] consists two parts: YF1 and FixJ. YF1 | + | We chose to use [http://parts.igem.org/Part:BBa_K592016 K592016] and [http://parts.igem.org/Part:BBa_K592006 K592006] as our light sensing device. [http://parts.igem.org/Part:BBa_K592016 K592016] consists two parts: YF1 and FixJ. YF1 is a blue-light sensor protein. It works in conjunction with its response regulator, FixJ. [http://parts.igem.org/Part:BBa_K592006 K592006] is a light-sensing promoter, which can express downstream gene when unphosphorylated FixJ presents. When exposed to blue-light, YF1 will be activated and phosphorylate FixJ. Phosphorylated FixJ cannot activate the light-sensing promoter [http://parts.igem.org/Part:BBa_K592006 K592006], and the expression of downstream gene will be repressed. |
| - | === | + | This picture can simply show the logic of this light-sensing device. |
| - | The | + | [[File:NYMU_light sensing 10-27.png|thumb|800px|center|Blue sensing device]] |
| - | + | ===Self-destruction Device=== | |
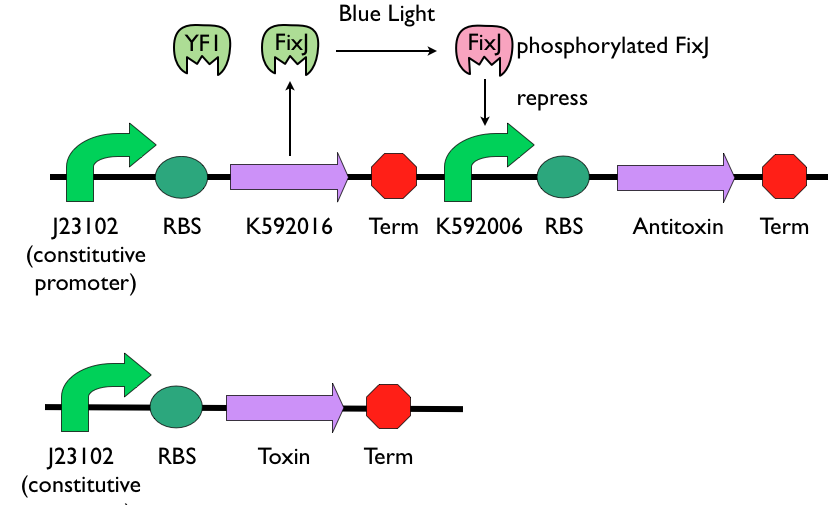
| + | The self-destruction device is composed of blue-light sensing promoter [http://parts.igem.org/Part:BBa_K592006 K592006], the blue-light sensing protein [http://parts.igem.org/Part:BBa_K592016 K592016], and a set of genes from [http://en.wikipedia.org/wiki/Toxin-antitoxin_system toxin-antitoxin system]. The toxin gene will be constitutively expressed. Antitoxin gene will be placed after [http://parts.igem.org/Part:BBa_K592006 K592006]. When toxin and antitoxin genes are both expressed, antitoxin can repress the function of toxin and the bacteria can survive. However, when toxin gene is dominating, it will kill the bacteria. | ||
| + | [[File:NYMU_self destruct device 10-27.png|thumb|800px|center|Self-destruction device]] | ||
==Circuit design and Experimental Method== | ==Circuit design and Experimental Method== | ||
===Circuit Design=== | ===Circuit Design=== | ||
| - | [http://parts.igem.org/Part:BBa_K592016 K592016] is cloned after constitutive promoter [http://parts.igem.org/Part:BBa_J23102 J23102], so the proteins YF1 and FixJ were continuing produced. When exposed to blue light, the inactive YF1 | + | [http://parts.igem.org/Part:BBa_K592016 K592016] is cloned after the constitutive promoter [http://parts.igem.org/Part:BBa_J23102 J23102], so the proteins YF1 and FixJ were continuing produced. Toxin gene is also cloned after [http://parts.igem.org/Part:BBa_J23102 J23102] and will be constitutively produced. When not exposed to blue light, the inactive YF1 will not phosphorylate FixJ, and the unphosphorylated FixJ can induce the downstream gene of promoter [http://parts.igem.org/Part:BBa_K592006 K592006], in this case, which is the antitoxin gene. When not exposed to blue-light, antitoxin can be expressed and consequently repress the function of toxin. When exposed to blue-light, the expression of antitoxin will be repressed, and constitutively produced toxin will kill ''Bee. coli''. |
| - | [[File: | + | [[File:NYMU_self desturct circuit off 10-27.png|thumb|800px|center|Without exposed to blue-light, Bee. coli survives]] |
| - | + | [[File:NYMU_self destruct circuit on 10-27.png|thumb|800px|center|Exposed to blue-light, Bee. coli will be killed]] | |
===Experimental Method=== | ===Experimental Method=== | ||
| - | First, we substitute | + | First, we substitute antitoxin with green fluorescent protein [http://parts.igem.org/Part:BBa_E0040 E0040] and replace toxin with red fluorescent protein [http://parts.igem.org/wiki/index.php?title=Part:BBa_E1010 E1010]. By calculating the ratio of GFP and RFP, we can derive the expression ratio of toxin and antitoxin. |
| - | [[File: | + | [[File:NYMU self destruct testing 10-27.png|thumb|800px|center|Testing Circuit]] |
Next, by comparing the number of colonies of the plate that is exposed to light and the plate that is blocked from light after 16 hours of incubation, we can characterize the functions of our device. | Next, by comparing the number of colonies of the plate that is exposed to light and the plate that is blocked from light after 16 hours of incubation, we can characterize the functions of our device. | ||
| - | [[File:NYMU_Light is block by aluminum foil.png|thumb|500px|center|'''Left: Light is | + | [[File:NYMU_Light is block by aluminum foil.png|thumb|500px|center|'''Left: Light is blocked by aluminum foil.'''<br><br>'''Right: Plate is exposed to light.''']] |
| + | This is the actual device we used to conduct this experiment. | ||
| + | [[File:NYMU light incubate device.jpg|thumb|800px|center|This device consists of two sets of blue-light LEDs]] | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 17:02, 27 October 2013


Contents |
Eco-friendly E. coli
Introduction
Our Bee. coli can express antimicrobial peptides such as defensin and abaecin to fight against Nosema cerenae. However, it is also possible that our Bee. coli can contaminate the natural environment and cause death to other species. Therefore, a light-induced lysis system was created to ensure our Bee. coli only lives inside of the bee.
Background
Blue Light Sensing Device
We chose to use [http://parts.igem.org/Part:BBa_K592016 K592016] and [http://parts.igem.org/Part:BBa_K592006 K592006] as our light sensing device. [http://parts.igem.org/Part:BBa_K592016 K592016] consists two parts: YF1 and FixJ. YF1 is a blue-light sensor protein. It works in conjunction with its response regulator, FixJ. [http://parts.igem.org/Part:BBa_K592006 K592006] is a light-sensing promoter, which can express downstream gene when unphosphorylated FixJ presents. When exposed to blue-light, YF1 will be activated and phosphorylate FixJ. Phosphorylated FixJ cannot activate the light-sensing promoter [http://parts.igem.org/Part:BBa_K592006 K592006], and the expression of downstream gene will be repressed. This picture can simply show the logic of this light-sensing device.
Self-destruction Device
The self-destruction device is composed of blue-light sensing promoter [http://parts.igem.org/Part:BBa_K592006 K592006], the blue-light sensing protein [http://parts.igem.org/Part:BBa_K592016 K592016], and a set of genes from [http://en.wikipedia.org/wiki/Toxin-antitoxin_system toxin-antitoxin system]. The toxin gene will be constitutively expressed. Antitoxin gene will be placed after [http://parts.igem.org/Part:BBa_K592006 K592006]. When toxin and antitoxin genes are both expressed, antitoxin can repress the function of toxin and the bacteria can survive. However, when toxin gene is dominating, it will kill the bacteria.
Circuit design and Experimental Method
Circuit Design
[http://parts.igem.org/Part:BBa_K592016 K592016] is cloned after the constitutive promoter [http://parts.igem.org/Part:BBa_J23102 J23102], so the proteins YF1 and FixJ were continuing produced. Toxin gene is also cloned after [http://parts.igem.org/Part:BBa_J23102 J23102] and will be constitutively produced. When not exposed to blue light, the inactive YF1 will not phosphorylate FixJ, and the unphosphorylated FixJ can induce the downstream gene of promoter [http://parts.igem.org/Part:BBa_K592006 K592006], in this case, which is the antitoxin gene. When not exposed to blue-light, antitoxin can be expressed and consequently repress the function of toxin. When exposed to blue-light, the expression of antitoxin will be repressed, and constitutively produced toxin will kill Bee. coli.
Experimental Method
First, we substitute antitoxin with green fluorescent protein [http://parts.igem.org/Part:BBa_E0040 E0040] and replace toxin with red fluorescent protein [http://parts.igem.org/wiki/index.php?title=Part:BBa_E1010 E1010]. By calculating the ratio of GFP and RFP, we can derive the expression ratio of toxin and antitoxin.
Next, by comparing the number of colonies of the plate that is exposed to light and the plate that is blocked from light after 16 hours of incubation, we can characterize the functions of our device.
This is the actual device we used to conduct this experiment.
 "
"