Team:Leeds/Project
From 2013.igem.org
m |
m |
||
| (91 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Team:Leeds_TouchGalleryCSS}} | |
| - | {{Team: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | }} | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
| - | < | + | <head profile="https://2013.igem.org/Team:Leeds/Project"> |
| - | < | + | <link rel="icon" |
| - | + | type="image/png" | |
| - | + | href="https://static.igem.org/mediawiki/2013/4/4d/Leeds_favicon.png"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <!-- | + | <!-- Javascript Gallery Code --> |
| + | <script type="text/javascript" src="https://2013.igem.org/Template:Team:Leeds_ajaxjs?action=raw&ctype=text/javascript"></script> | ||
| + | <script type="text/javascript" src="https://2013.igem.org/Template:Team:Leeds_TouchSwipejs?action=raw&ctype=text/javascript"></script> | ||
| + | <script src="https://2013.igem.org/Template:Team:Leeds_TouchGalleryjs?action=raw&ctype=text/javascript"> | ||
| - | + | /*********************************************** | |
| - | + | * Touch Image Gallery- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) | |
| - | + | * This notice MUST stay intact for legal use | |
| - | + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for this script and 100s more | |
| - | + | ***********************************************/ | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | </script> | ||
| + | <script> | ||
| - | + | jQuery(function(){ // on DOM load | |
| + | $('div#touchgallery1').touchgallery({ // initialize first demo | ||
| + | width: 750, | ||
| + | height: 500 | ||
| + | }) | ||
| + | $('div#touchgallery2').touchgallery({ // initialize second demo | ||
| + | width: 400, | ||
| + | height: 350 | ||
| + | }) | ||
| + | }) | ||
| - | + | </script> | |
| - | + | </head> | |
| - | + | </html> | |
| - | + | __NOTOC__ | |
| - | + | {{Team:Leeds_layout|HeaderParky=</div>[[File:Leeds_ParkytopProject.png|150px|link=|frameless]]<div>|HeaderImage=[[File:Leeds_ProjectHead.png|825px|link=|frameless]]|Header=MicroBeagle|content= | |
| - | + | <div><div id="touchgallery1" class="touchgallery"> | |
| - | + | <ul> | |
| - | + | <li>[[File:Leeds_Microbeagleheader.png|700px|center|Beagles vs microBeagles|frameless]]</li> | |
| - | == | + | <li>[[File:Leeds_bb1schematic.png|700px|center|Our first biosystem will be used to help characterise the Cpx pathway, which reacts to membrane stress|frameless]]</li> |
| - | + | <li>[[File:Leeds_Device2schematic.png|700px|center|Our second biosystem will test the INP protein's capability to express any protein we like on the surface, in this case, a silica binding protein|frameless]]</li> | |
| - | + | </ul> | |
| - | + | </div></div> | |
| - | + | <br> | |
| - | + | <h2><font size="6">'''Our Project'''</font></h2> | |
| - | === | + | ==What is MicroBeagle I hear you cry?== |
| - | + | Well, MicroBeagle is an ''E.coli'' cell that has been modified so that it can help “hunt down” pathogens. Just like a Beagle, the hunting dog, searches for, finds, and indicates the presence of rabbits and hares, our MircoBeagle finds and points to pathogens, detecting them through physical binding, and lighting up to indicate their presence! By utilising the Cpx pathway, which responds to membrane stress (which we hypothesize would be induced by the binding event) and GFP (as a reporter) we hope to create a new biosensor that ultimately will give diagnostic results that can be seen with the naked eye. | |
| - | + | <br style="clear:both"/> | |
| - | + | [[File:PhysicalBinding.png|left|300px|We are using Physical binding!|link=|frameless]] | |
| - | + | Our MicroBeagle project is laying groundwork towards the development of a modular, general platform for pathogen detection. What makes Leeds iGEM's MicroBeagle distinctive to previous strategies used for analyte detection within iGEM is that we use physical binding as our detection mode, versus sensing of soluble analytes as has been explored in previous iGEM projects. A physical binding detection mode should be well suited to assaying for a wide range of pathogen targets, as tailored target-binding proteins or peptides can be displayed from the cell surface, allowing specific binding to the pathogen of interest. | |
| - | + | <br> | |
| - | === | + | <br style="clear:both"/> |
| - | + | ==Why is the MircoBeagle a useful device?== | |
| - | + | There could be many different possible uses for our device, as the pathogen detection mechanism we are using is modular and can easily be tailored to individual needs. This is achieved by swapping out the gene coding the cell surface-displayed binding domain to suit your needs. We envision the MircoBeagle being used for testing blood samples, detection of heavy metals and, in the application that is our main inspiration, detection of pathogens in water samples. | |
| - | + | <br> | |
| - | + | ==Why Water Testing is so important== | |
| - | === | + | [[File:Dirtywater.png|right|300px|Would you like to have to drink this water?|link=|frameless]] |
| - | + | Detecting pathogens in water is a really important application as 3.4 million people a year die from water related diseases<sup>[http://www.who.int/water_sanitation_health/takingcharge.html [1]],[http://water.org/water-crisis/water-facts/water/ [2]]</sup>. | |
| - | + | <br> | |
| - | + | 768 million people in the world do not have access to safe water<sup>[http://www.unicefusa.org/work/water/ [3]]</sup>. This is roughly one in ten of the world's population, the majority of which is caused by faecal contamination: poor sewage systems that flow back into the water supply<sup>[http://www.who.int/water_sanitation_health/bathing/srwe1-chap4.pdf [4]]</sup>. | |
| - | + | <br> | |
| - | == | + | The issue is actually more complex and subtle than just direct health problems, as the knock on economic problems that result from poor sanitation are also damaging. Lack of water, sanitation and hygiene costs Sub-Saharan African countries more in lost GDP than the entire continent gets in development aid<sup>[http://dspace.cigilibrary.org/jspui/bitstream/123456789/20135/1/Human%20Development%20Report%202006%20Beyond%20Scarcity%20Power%20poverty%20and%20the%20global%20water%20crisis.pdf?1 [5]]</sup>. The figures are shocking and we as a team want to create something that may, in the future, help to reduce those numbers. |
| + | <br> | ||
| + | While many organizations are creating new, appropriate water purification devices and systems, we realize these will only be as good as the measurement technologies available to test their performance will allow. This measurement bottleneck is particularly critical in infrastructure-poor areas with no or limited access to the molecular biology labs typically used to assay for water-borne pathogens. | ||
| + | <br> | ||
| + | While many teams around the world are seeking new ways of transporting chassis and bio-devices in facile, safe, and reliable manners (e.g., through capsules or spores), we felt that efforts to develop modular pathogen detection systems were lacking. Once effective transport strategies are established, we hope to have a detection system ready-to-go for on-site water testing and diagnostics. | ||
| + | <br> | ||
| + | To address this issue using a novel synthetic biological approach, the MircoBeagle was born! | ||
| + | <br> | ||
| + | ===Below is a summary of our Device development strategy...=== | ||
| + | ==Phase I== | ||
| + | We split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within iGEM and beyond. Our two devices include a device for activating a reporter in response to membrane stress (Device 1) and a device for binding to a physical target at the cell surface and thereby activate the membrane-stress-reporter (Device 2). | ||
| + | ===Device 1=== | ||
| + | [[File:Leeds_bb1schematic.png|left|400px|Cartoon schematic of Biosystem 1|link=|frameless]] | ||
| + | Device 1 is a very simple modification of the pCpxR promoter in ''E. coli'' to produce a fluorescent reporter when the cell is under membrane stress. Activation of Cpx pathways in response to solid surface interactions was first reported by [http://www.ncbi.nlm.nih.gov/pubmed/11830644 ''Otto & Silhaevy 2001''] in which hydrophobically treated glass beads were used to induce membrane stress. Given that the promoter pCpxR was specifically shown to be activated by this interaction, our hope is that, in initial stages of MicroBeagle development, this promoter can respond to a wide variety of physical binding interactions when these are specifically designed for target surfaces. pCpxR is present in the registry of standard biological parts, examined previously by [http://parts.igem.org/wiki/index.php?title=Part:BBa_K339007 Calgary 2010] and [http://parts.igem.org/Part:BBa_K135001?title=Part:BBa_K135001 BCCS-Bristol 2008]. In our MicroBeagle work, we aim to further explore and characterize this promoter, and investigate for the first time the utility of this promoter in generating a reporting signal when specifically coupled with a designed cell surface-solid target binding interaction. We characterized our Device 1, pCpxR-GFP, by testing fluorescence generation vs a variety of potential membrane-stress-inducing environments, including various concentrations of a detergent (triton X-100) and the presence of various solid particles. | ||
| + | <br style="clear:both"/> | ||
| + | ===Si4 Binding Peptide=== | ||
| + | Our second device of interest ("Device 2", described below) utilizes, in addition to the pCpxR reporter system (above), a surface-displayed target-binding moiety for physical attachment to particles. Display of the target-binding moiety utilizes Ice Nucleation Protein (INP) to display an oligo-peptide of our choice on the outer-membrane of our ''E. coli''. Initially this will be a peptide capable of binding silica beads, allowing us to create a non-toxic model system of pathogen detection for initial stages of device development. INP is a transmembrane protein that expresses any sequence that is placed on the C-terminus of its gene on the outer surface of the cell. Specifically, we utilized the INP display system developed by [http://parts.igem.org/Part:BBa_K811003?title=Part:BBa_K811003 UPenn 2012]. The silica binding domain we utilized, termed "Si4" was developed by [http://www.ingentaconnect.com/content/asp/jnn/2002/00000002/00000001/art00015 Naik, Lawrence, Clarson, and Stone 2002]. This silica-binding polypeptide was discovered via phage display screening, and was also found to precipitate silica when reacted with silicic acid. This provided us with an additional route to test for the presence of Si4 on cell surfaces, i.e. via mineralization assays using silicic acid as a mineral precursor. | ||
| + | <br style="clear:both"/> | ||
| + | <br> | ||
| + | ==Phase II== | ||
| + | Phase II takes the products of Phase I to the next level, and begins to look at integration of the INP-displayed Si4 binding peptide and Device 1 to give a new device; Device 2. | ||
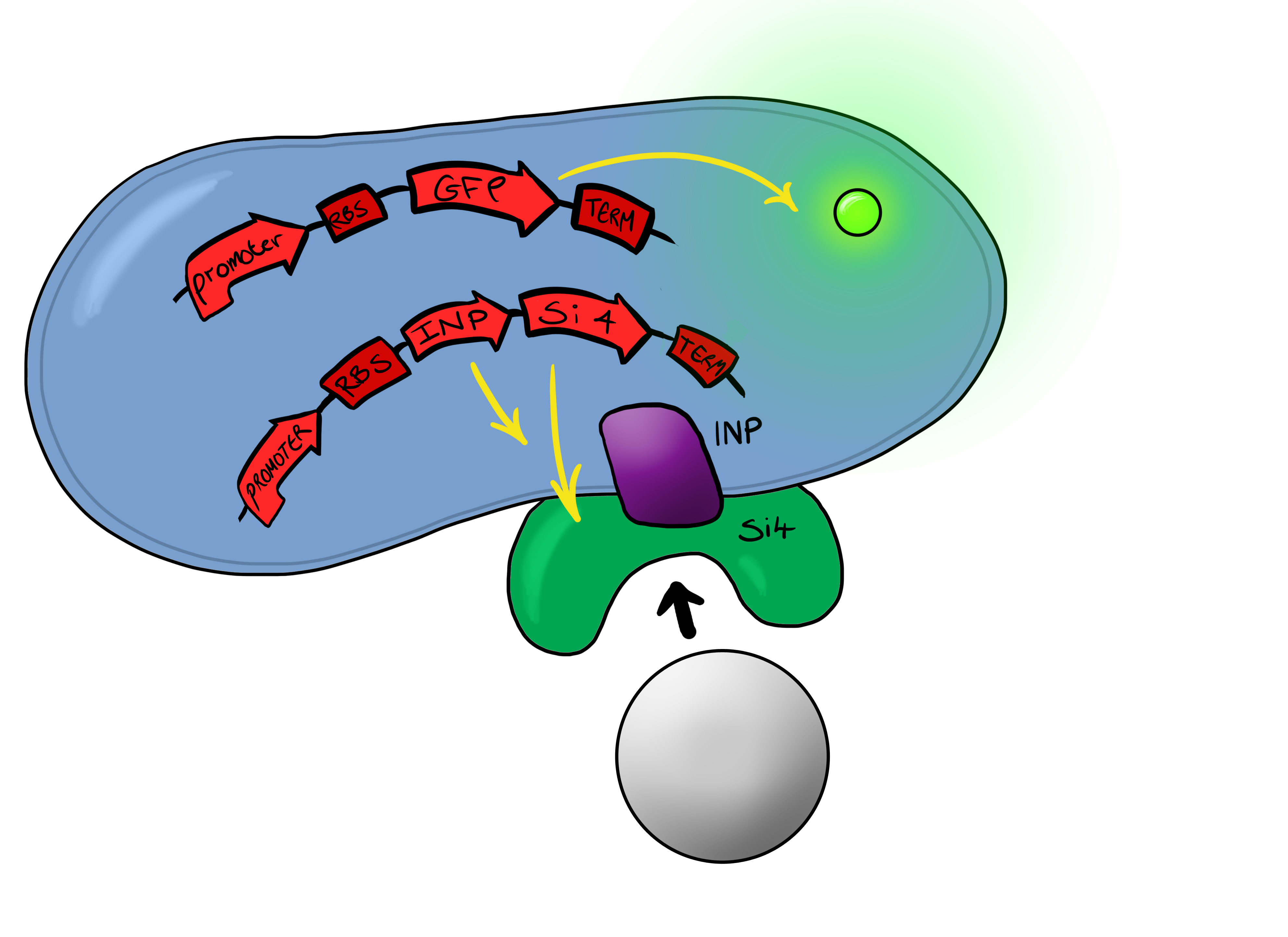
| + | ===Device 2=== | ||
| + | [[File:Biosystem3.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | ||
| + | Device 2 brings together on the same plasmid the genes that code for (1) the CpxP promoter controlling GFP expression and (2) the INP-displayed Si4 binding domain. The effect of this is that when the Si4 binding domain binds to the silica target, surface stress propagated through the INP protein will activate the pCpxR promoter through the Cpx signaling pathway. This will cause GFP to be produced and thereby act as a visible signal that the 'pathogen' has been detected. | ||
| + | <br style="clear:both"/> | ||
| + | This is the final MircoBeagle device and has the potential to be made into many different pathogen detectors. This can easily be done by swapping out the gene coding for the binding moiety next to the INP gene. Once Device 2 has been suitably characterised and all bugs fixed, the next step will be to swap the silca binding domain for a binding domain that binds to a particular pathogen. | ||
| + | <br> | ||
| + | We note that given the conceptual modularity of our MicroBeagle strategy, in principle any of the key device elements--surface-displayed binding domain, transmembrane scaffold protein, stress-activated promoter, and/or fluorescent or colorimetric report--can be optimized or exchanged for new specific elements that exhibit the intended component function. In the future, performance of the device components should be amenable to evolutionary improvement, either individually or in tandem. In fact, the binding domain element would benefit from a plethora of evolutionary screens that have already been conducted in biomedical and biotechnological labs worldwide to identify protein and peptide binders to a wide range of both biological and non-biological targets. | ||
| + | <br style="clear:both"/> | ||
| + | ==The Cpx Pathway== | ||
| + | This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | ||
| + | [[File:CpxPathway.png|center|600px|Cartoon schematic of the Cpx Pathway|link=|frameless]] | ||
| + | <br style="clear:both"/> | ||
| + | ===A brief explanation of the Cpx Pathway=== | ||
| + | The Cpx pathway responds to various membrane stresses; it is a two component signal transduction pathway. The first component is CpxA, an inner membrane protein which, when bound to CpxP (an inhibitor molecule), is inactive. When CpxP is unbound from CpxA, CpxA undergoes autophosphorylation. A kinase enzyme then phosphorylates CpxR, the second component in the pathway, which then binds to the pCpxR promoter and activates transcription.<br> | ||
| + | There is still a lot of debate regarding the detailed mechanisms of the pathway, with many different theories proposed, but it is thought that membrane stress causes the misfolding of proteins (pili being the main contributor), which in turn causes misfolded subunits to accumulate in the periplasm, which in turn titrates the CpxP inhibitor molecule into the unbound state, thereby activating the pathway. We have started development of a [[Team:Leeds/Modeling|rules-based model]] to help discern the processes involved with the pathway. | ||
| + | }} | ||
Latest revision as of 02:03, 5 October 2013
|
Our ProjectWhat is MicroBeagle I hear you cry?Well, MicroBeagle is an E.coli cell that has been modified so that it can help “hunt down” pathogens. Just like a Beagle, the hunting dog, searches for, finds, and indicates the presence of rabbits and hares, our MircoBeagle finds and points to pathogens, detecting them through physical binding, and lighting up to indicate their presence! By utilising the Cpx pathway, which responds to membrane stress (which we hypothesize would be induced by the binding event) and GFP (as a reporter) we hope to create a new biosensor that ultimately will give diagnostic results that can be seen with the naked eye.
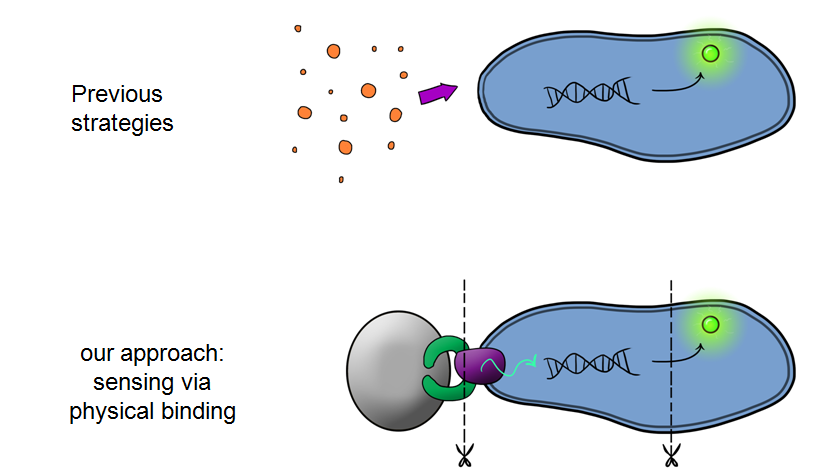 Our MicroBeagle project is laying groundwork towards the development of a modular, general platform for pathogen detection. What makes Leeds iGEM's MicroBeagle distinctive to previous strategies used for analyte detection within iGEM is that we use physical binding as our detection mode, versus sensing of soluble analytes as has been explored in previous iGEM projects. A physical binding detection mode should be well suited to assaying for a wide range of pathogen targets, as tailored target-binding proteins or peptides can be displayed from the cell surface, allowing specific binding to the pathogen of interest.
Why is the MircoBeagle a useful device?There could be many different possible uses for our device, as the pathogen detection mechanism we are using is modular and can easily be tailored to individual needs. This is achieved by swapping out the gene coding the cell surface-displayed binding domain to suit your needs. We envision the MircoBeagle being used for testing blood samples, detection of heavy metals and, in the application that is our main inspiration, detection of pathogens in water samples.
Why Water Testing is so important Detecting pathogens in water is a really important application as 3.4 million people a year die from water related diseases[http://www.who.int/water_sanitation_health/takingcharge.html [1]],[http://water.org/water-crisis/water-facts/water/ [2]].
Below is a summary of our Device development strategy...Phase IWe split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within iGEM and beyond. Our two devices include a device for activating a reporter in response to membrane stress (Device 1) and a device for binding to a physical target at the cell surface and thereby activate the membrane-stress-reporter (Device 2). Device 1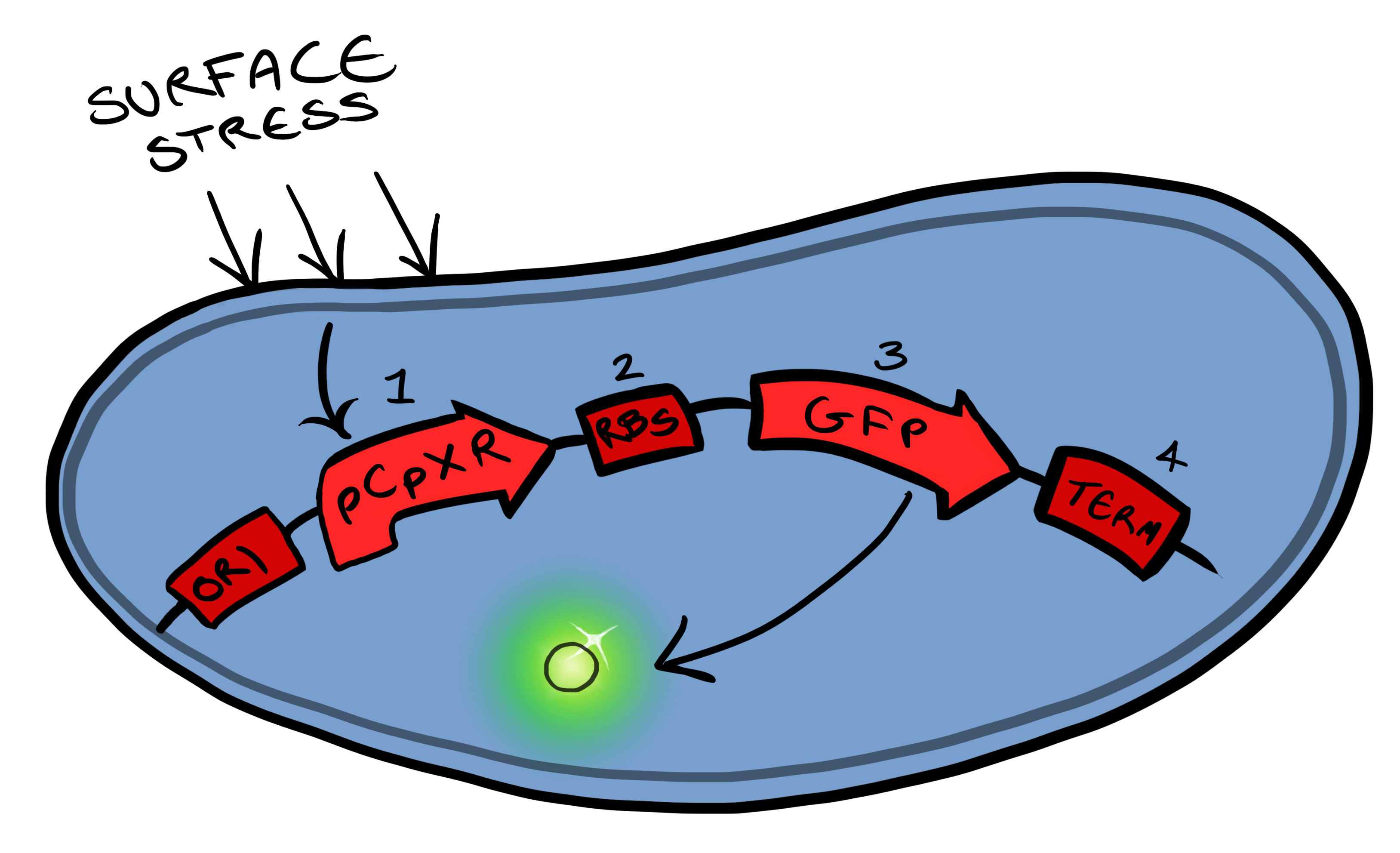 Device 1 is a very simple modification of the pCpxR promoter in E. coli to produce a fluorescent reporter when the cell is under membrane stress. Activation of Cpx pathways in response to solid surface interactions was first reported by [http://www.ncbi.nlm.nih.gov/pubmed/11830644 Otto & Silhaevy 2001] in which hydrophobically treated glass beads were used to induce membrane stress. Given that the promoter pCpxR was specifically shown to be activated by this interaction, our hope is that, in initial stages of MicroBeagle development, this promoter can respond to a wide variety of physical binding interactions when these are specifically designed for target surfaces. pCpxR is present in the registry of standard biological parts, examined previously by [http://parts.igem.org/wiki/index.php?title=Part:BBa_K339007 Calgary 2010] and [http://parts.igem.org/Part:BBa_K135001?title=Part:BBa_K135001 BCCS-Bristol 2008]. In our MicroBeagle work, we aim to further explore and characterize this promoter, and investigate for the first time the utility of this promoter in generating a reporting signal when specifically coupled with a designed cell surface-solid target binding interaction. We characterized our Device 1, pCpxR-GFP, by testing fluorescence generation vs a variety of potential membrane-stress-inducing environments, including various concentrations of a detergent (triton X-100) and the presence of various solid particles.
Si4 Binding PeptideOur second device of interest ("Device 2", described below) utilizes, in addition to the pCpxR reporter system (above), a surface-displayed target-binding moiety for physical attachment to particles. Display of the target-binding moiety utilizes Ice Nucleation Protein (INP) to display an oligo-peptide of our choice on the outer-membrane of our E. coli. Initially this will be a peptide capable of binding silica beads, allowing us to create a non-toxic model system of pathogen detection for initial stages of device development. INP is a transmembrane protein that expresses any sequence that is placed on the C-terminus of its gene on the outer surface of the cell. Specifically, we utilized the INP display system developed by [http://parts.igem.org/Part:BBa_K811003?title=Part:BBa_K811003 UPenn 2012]. The silica binding domain we utilized, termed "Si4" was developed by [http://www.ingentaconnect.com/content/asp/jnn/2002/00000002/00000001/art00015 Naik, Lawrence, Clarson, and Stone 2002]. This silica-binding polypeptide was discovered via phage display screening, and was also found to precipitate silica when reacted with silicic acid. This provided us with an additional route to test for the presence of Si4 on cell surfaces, i.e. via mineralization assays using silicic acid as a mineral precursor.
Phase IIPhase II takes the products of Phase I to the next level, and begins to look at integration of the INP-displayed Si4 binding peptide and Device 1 to give a new device; Device 2. Device 2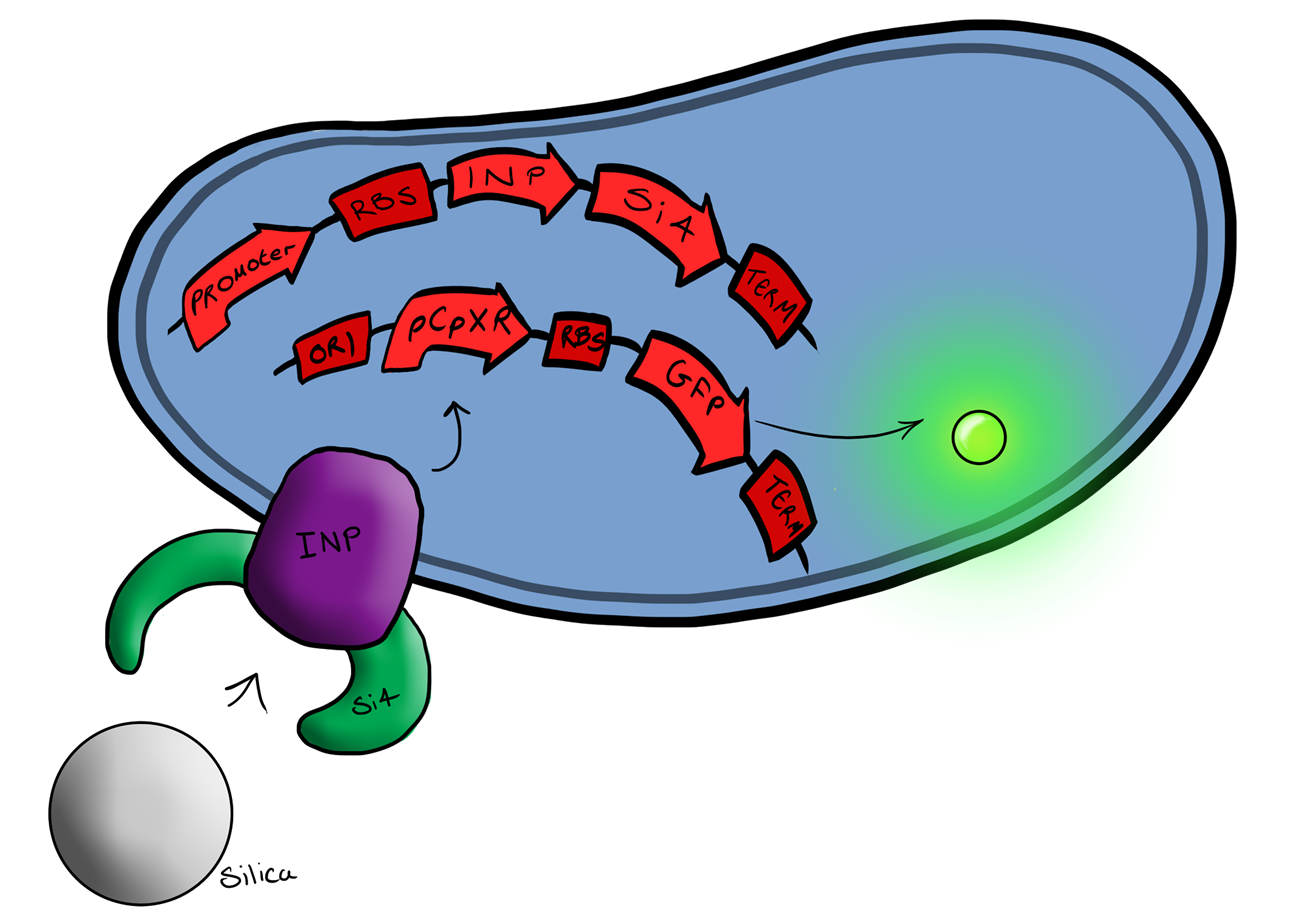 Device 2 brings together on the same plasmid the genes that code for (1) the CpxP promoter controlling GFP expression and (2) the INP-displayed Si4 binding domain. The effect of this is that when the Si4 binding domain binds to the silica target, surface stress propagated through the INP protein will activate the pCpxR promoter through the Cpx signaling pathway. This will cause GFP to be produced and thereby act as a visible signal that the 'pathogen' has been detected.
The Cpx PathwayThis project is highly dependent upon a naturally occuring pathway in E.Coli called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. 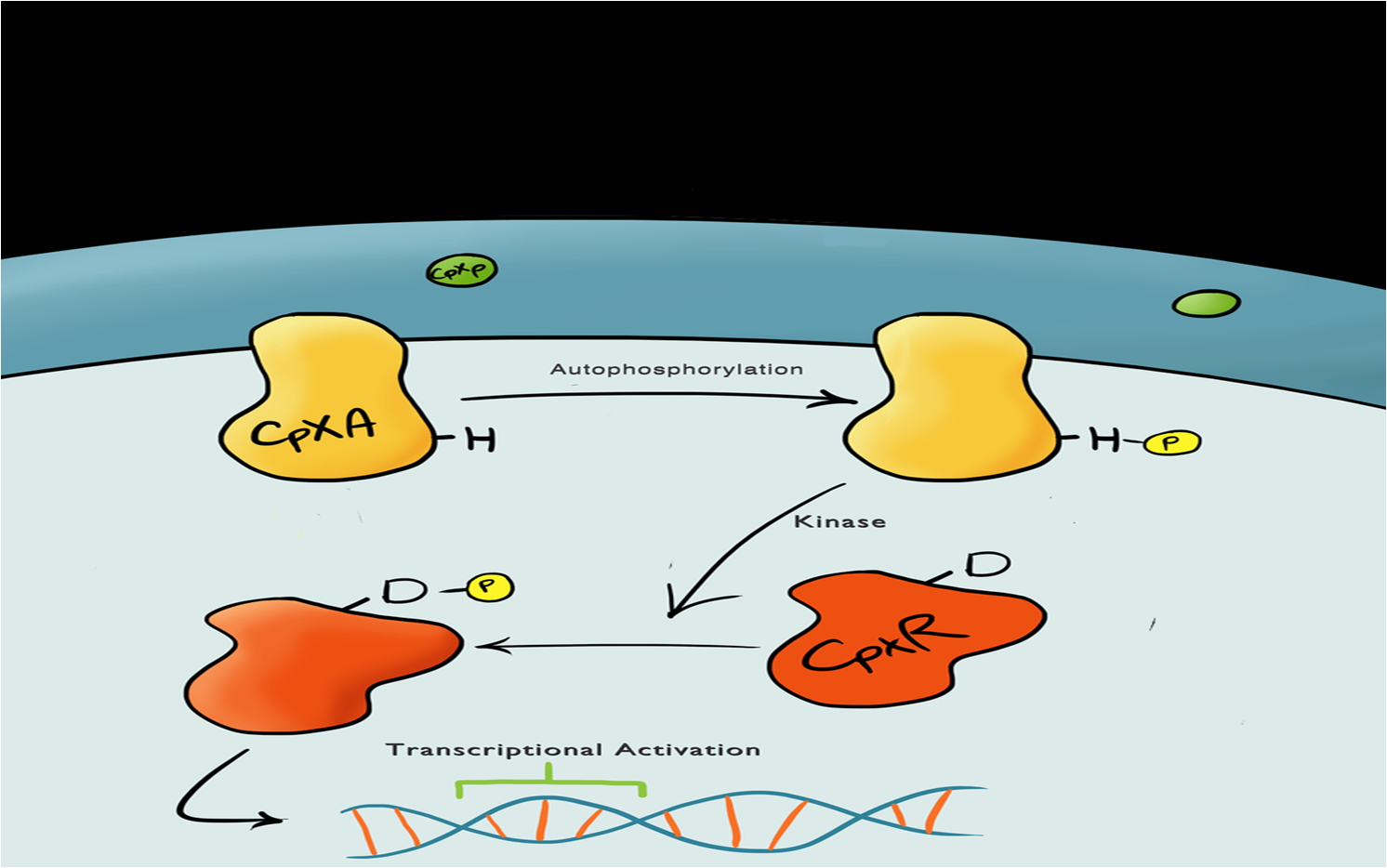
A brief explanation of the Cpx PathwayThe Cpx pathway responds to various membrane stresses; it is a two component signal transduction pathway. The first component is CpxA, an inner membrane protein which, when bound to CpxP (an inhibitor molecule), is inactive. When CpxP is unbound from CpxA, CpxA undergoes autophosphorylation. A kinase enzyme then phosphorylates CpxR, the second component in the pathway, which then binds to the pCpxR promoter and activates transcription. | |||||||
 |
| ||||||

| |||||||

| |||||||
 "
"