Team:INSA Toulouse/contenu/lab practice/results/polT7
From 2013.igem.org
| (One intermediate revision not shown) | |||
| Line 164: | Line 164: | ||
<h2 class="title2">Result</h2> | <h2 class="title2">Result</h2> | ||
| - | <p class="texte">The addition of a promoter and a rbs to the T7 polymerase gene was successful. However, we did not have time to go further and add the terminator to the construct. Nevertheless, a new biobrick pT7-RFP was created! | + | <p class="texte">The addition of a promoter and a rbs to the T7 polymerase gene was successful. However, we did not have time to go further and add the terminator to the construct.<br> Nevertheless, a new biobrick pT7-RFP was created!<br> |
Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction with EcoRI/PstI:<br><br> | Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction with EcoRI/PstI:<br><br> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/f/f9/Result_polT7_2.jpg" class=" | + | <img src="https://static.igem.org/mediawiki/2013/f/f9/Result_polT7_2.jpg" class="imgcenter" /><br><br> |
Verifications of the validity of the constructions were performed with visual inspection of the colours of the colonies. The pT7-RFP plasmid was transformed into competent BL21-DE3. Plates were supplemented or not with IPTG. White colonies were obtained on the Petri dishes with no IPTG and red colonies on the Petri dishes containing IPTG. </p> | Verifications of the validity of the constructions were performed with visual inspection of the colours of the colonies. The pT7-RFP plasmid was transformed into competent BL21-DE3. Plates were supplemented or not with IPTG. White colonies were obtained on the Petri dishes with no IPTG and red colonies on the Petri dishes containing IPTG. </p> | ||
Latest revision as of 15:13, 4 October 2013
Results - T7 Polymerase characterization
Objective
Characterize the activity of T7 polymerase with an RFP reporter under a T7 promoter. Reminder: T7 polymerase is necessary in our system to activate the two T7 promoters in the AND1 and AND2 gate.
Conception
The following constructions were designed:
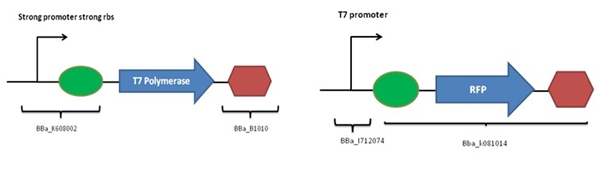
The coding sequence of T7 polymerase was extracted from the genome of E. coli BL21-DE3. Further cloning steps were necessary to add a promoter, a rbs and a terminator. The expected functioning is:
- Production of RFP if T7 polymerase is present (red colonies)
- Absence of RFP if T7 polymerase is absent (white colonies)
Result
The addition of a promoter and a rbs to the T7 polymerase gene was successful. However, we did not have time to go further and add the terminator to the construct.
Nevertheless, a new biobrick pT7-RFP was created!
Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction with EcoRI/PstI:

Verifications of the validity of the constructions were performed with visual inspection of the colours of the colonies. The pT7-RFP plasmid was transformed into competent BL21-DE3. Plates were supplemented or not with IPTG. White colonies were obtained on the Petri dishes with no IPTG and red colonies on the Petri dishes containing IPTG.

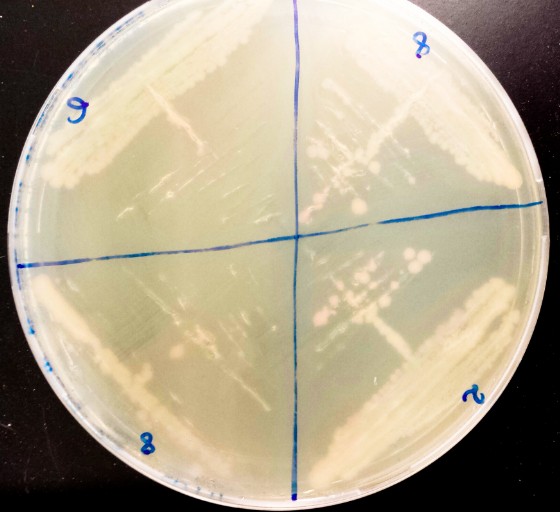
Discussion
The biobrick behave as expected. It was submitted to the registry as BBa_K1132045.
 "
"

