Team:TU-Delft/Peptides
From 2013.igem.org
| (9 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
| - | <div style="margin-left:30px;margin-right:30px; width:900px;float:left;"> | + | <div style="margin-left:30px;margin-right:30px;margin-top:-30px; width:900px;float:left;"> |
| Line 145: | Line 145: | ||
</p> | </p> | ||
| - | <p align="justify"> | + | <p align="justify"> A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter and the Ulp-1 production was driven by arabinose inducible promoter pBad. The plasmid was transformed into an BL21(DE3) pLysS strain. This construct was designed to check whether in vivo cleavage is possible. The main idea of the experiment is to first produce large amount of soluble fraction of SUMO-peptides and the produce the Ulp protease to cleave the sufficiently produced fusion peptides. </p> |
| - | A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter and the Ulp-1 production was driven by arabinose inducible promoter pBad. The plasmid was transformed into an BL21(DE3) pLysS strain. This construct was designed to check whether in vivo cleavage is possible. The main idea of the experiment is to first produce large amount of soluble fraction of SUMO-peptides and the produce the Ulp protease to cleave the sufficiently produced fusion peptides. | + | |
| - | </p> | + | |
<center> | <center> | ||
| Line 159: | Line 157: | ||
<h4>Results</h4> | <h4>Results</h4> | ||
| - | <p align="justify"> | + | <p align="justify"> The presence of plasmid with gene inserts encoding the SUMO-peptide (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022116" target="blank">BBa_K1022116</a>, <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022117" target="blank">BBa_K1022117</a>,<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022118" target="blank">BBa_K1022118</a>) was confirmed by a colony pcr. The expected size of the insert was approximately around ~1500 bp which was clearly evident from the agarose gel picture figure(<a href="https://2013.igem.org/Team:TU-Delft/Peptides#ulp1">9,10,11</a>). Though the actual size of the inserts are ~1300 bp the use of sequencing primers VF2 and VR which bind 100-150 base pair away from suffix and prefix the bands are around 1500 bp. An Eurogentec Smartladder MW-1700-10 [lane 1] (<a href="https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf" target="blank">https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf</a>) was used to identify the size of the fragments.</p> |
| - | The presence of plasmid with gene inserts encoding the SUMO-peptide (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022116" target="blank">BBa_K1022116</a>, <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022117" target="blank">BBa_K1022117</a>,<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022118" target="blank">BBa_K1022118</a>) was confirmed by a colony pcr. The expected size of the insert was approximately around ~1500 bp which was clearly evident from the agarose gel picture figure(<a href="https://2013.igem.org/Team:TU-Delft/Peptides#ulp1">9,10,11</a>). Though the actual size of the inserts are ~1300 bp the use of sequencing primers VF2 and VR which bind 100-150 base pair away from suffix and prefix the bands are around 1500 bp. An Eurogentec Smartladder MW-1700-10 [lane 1] (<a href="https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf" target="blank">https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf</a>) was used to identify the size of the fragments. | + | |
| - | </p> | + | |
| Line 170: | Line 166: | ||
</center> | </center> | ||
<center> | <center> | ||
| - | <p>Figure 9: pBad Ulp SUMO maximin-H5 Colony PCR <br><br></p></div></center> | + | <p>Figure 9: pBad Ulp SUMO maximin-H5 (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022116" target="blank">BBa_K1022116</a>) Colony PCR <br><br></p></div></center> |
<a name="picture"></a> | <a name="picture"></a> | ||
| Line 178: | Line 174: | ||
</center> | </center> | ||
<center> | <center> | ||
| - | <p>Figure 10:pBad Ulp SUMO magainin colony PCR <br><br></p></div></center> | + | <p>Figure 10:pBad Ulp SUMO magainin (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022118" target="blank">BBa_K1022118</a>) colony PCR <br><br></p></div></center> |
| Line 187: | Line 183: | ||
</center> | </center> | ||
<center> | <center> | ||
| - | <p>Figure 11:pBad Ulp SUMO signiferin colony PCR <br><br></p></div> | + | <p>Figure 11:pBad Ulp SUMO signiferin (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1022117" target="blank">BBa_K1022117</a>) colony PCR <br><br></p></div> |
</center> | </center> | ||
| Line 202: | Line 198: | ||
</center> | </center> | ||
<center> | <center> | ||
| - | <p>Figure 12:Coverage for SUMO without Peptide from Tandem MS <br><br></p></center> | + | <p>Figure 12:Coverage for SUMO without Peptide from Tandem MS [Red Letters] <br><br></p></center> |
<h4>Future Insights</h4> | <h4>Future Insights</h4> | ||
<p align="justify"> | <p align="justify"> | ||
| - | Some intense literature studies gave us some insights of how we can improve the design of the peptides. Usually, the frogs and toads have the peptides with some post translational modifications (PTM) like amidation <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references"> [5]</a>,polyglycylation <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references"> [6]</a> ,etc. These make them less vulnerable to carboxypeptidases or amidopeptidases. But, the major hurdle is that the E. coli cannot perform these PTMs that made us choose peptides without PTMs. This makes it clear that there are more possibilities that the peptides could be cleave of by the proteases. | + | Some intense literature studies gave us some insights of how we can improve the design of the peptides. Usually, the frogs and toads have the peptides with some post translational modifications (PTM) like amidation <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references"> [5]</a>,polyglycylation <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references"> [6]</a> ,etc. These make them less vulnerable to carboxypeptidases or amidopeptidases. But, the major hurdle is that the <i> E. coli</i> cannot perform these PTMs that made us choose peptides without PTMs. This makes it clear that there are more possibilities that the peptides could be cleave of by the proteases. |
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
| - | We also found that some of the antimicrobial peptide (AMP) group called bombinins <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references">[7]</a> was similar with maximins, which by itself is from the <i>Bombina</i> species. But, the bombinins have D-isoleucine instead of L-isoleucine. This makes it difficult for the peptidases to cleave the peptides. So, the maximins could also have some D-aminoacids that makes it much more difficult to perform these PTMs using E. coli. | + | We also found that some of the antimicrobial peptide (AMP) group called bombinins <a href="https://2013.igem.org/Team:TU-Delft/Peptides#references">[7]</a> was similar with maximins, which by itself is from the <i>Bombina</i> species. But, the bombinins have D-isoleucine instead of L-isoleucine. This makes it difficult for the peptidases to cleave the peptides. So, the maximins could also have some D-aminoacids that makes it much more difficult to perform these PTMs using <i> E. coli</i>. </p> |
<p align="justify"> | <p align="justify"> | ||
Thus, in near future these peptides can be made if we can manage to make create a strain that can perform these PTMs so we can use them as a chassis to produce our peptides | Thus, in near future these peptides can be made if we can manage to make create a strain that can perform these PTMs so we can use them as a chassis to produce our peptides | ||
Latest revision as of 21:17, 4 October 2013

Peptide production
The era of antibiotics are now outdated as the microorganisms started gaining new resistance mechanism to evade the effects of antibiotics. Nowadays more and more researches are focused towards new antimicrobial that can be more effective against these pathogens.
The antimicrobial peptides (AMP), natural or de-novo are becoming more popular due to their potential to kill or restrict the growth of microorganisms. They form a part of chemical defense against predators. All organisms, including higher order eukaryotes have some antimicrobial peptides that acts against specific strains of microorganisms.They are usually charged and interact with the opposite charges present on the membrane thus boring a hole through the membrane subsequently lysing the cell [1].Unlike antibiotics, most AMPs do not interfere in the genetic, transcriptional or translational machinery of the cell.
We chose 3 AMP that are produced in the skins of toads and frogs. They are magainin (GIGKFLHSAKKFGKAFVGQIMNS) from Xenopus laevis , signiferin (IIGHLIKTALGMLGL) from Crinia signifera and maximin-H5 (ILGPVLGLVSDTLDDVLGIL) from Bombina maxima. The reasons for choosing these AMP are because of their target specific nature, charge and length. The peptides are described in literature as being specifically toxic to certain species, in this case a low MIC for Staphylococci. The widely accepted theory behind this selectivity is the idea the peptides have a localized charge in either their C- or N-terminal part. This charge is needed to interact with the membrane of the cell before they can bore holes in them. Another important criterion used in selection of the peptides has been the published toxicity towards mammalian cells with human cells in specia, which is important in making them possible candidates as pharmaceuticals [2].
The recombinant production of these peptides in E.coli would help large scale production of these peptides and administer them as antimicrobials [5]. But, the major hurdle in producing them in vivo is the formation of inclusion bodies as they are charged. This could be overcome by making protein fusion with SUMO (Small Ubiquitin like Modifier), MBP (Maltose Binding Protein), Trx (Thioredoxin) etc.
In this project, a SUMO-peptide fusion was opted [3] as a suitable expression system as they make the fusion proteins more soluble. The Ulp-1 cleaving the fusion is a special protease that can cleave of the fusion without leaving any scar. This protease is also expressed and controlled by a negative transcriptional cascade. As the fusion is produced it triggers a cascade that ultimately leads to the expression of Ulp-1 and facilitates in vivo cleavage. This fusion increases the solubility of the peptide and reduces inclusion bodies formation.(Figure 1).

Figure 1: Schematic diagram of the peptide production
SUMO-Peptide Production
Aim: Increasing solubility of peptide with Sumo Fusion.
Description
The peptide by itself is not soluble in the cytoplasm but making a fusion of peptide with Small Ubiquitin like Modifiers (SUMO) will increase the solubility of the peptide, thus increasing the cytoplasmic fraction of the peptide.
A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter. This gene containing plasmid was harboured in a BL21(DE3) strain that has lac promoter driven T7 polymerase. Upon induction by IPTG the SUMO peptide fusion is produced as a soluble protein fraction.
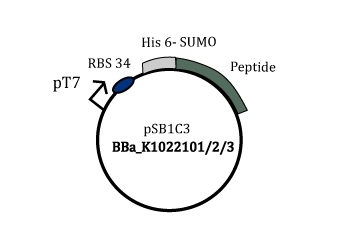
Figure 2: Part BBa_K1022101/2/3
Results
The presence of plasmid with gene inserts encoding the SUMO-peptide (BBa_K1022101, BBa_K1022102,BBa_K1022103) was confirmed by a colony pcr. The expected size of the insert with the T7 promoter was approximately around 400 bp which was clearly evident from the agarose gel picture Figure 3. An Eurogentec Smartladder MW-1700-10 (https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf) was used to identify the size of the fragments.

Figure 3:Colony PCR SUMO Peptide
Then strains were subjected to protein expression by means of IPTG induction. The cell lysate and the centrifuged cell debris were run on a 16 % tricine gels at 90 V for 90 minutes. The figure 4 clearly shows a over-expression of our fusion protein at ~15 kDa both in whole cell lysate (CL) and the membrane (M), This was from just 10 µg of total protein for cell lysate (CL) and 30 µg of total protein in the membrane (M).
Then just the induced CL and M were run with 30 µg of total protein per well as in figure 5 the results were again consistent (Black circle). One of the over-expressed band was excised and subjected for in-gel tryptic digestion and then to MS/MS measurement in Q-trap MS instrument (Thermo Scientific). To prove the solubility of the peptide the the CL and M from constructs that express peptide without SUMO was run with the CL from SUMO- peptide as in figure 6.

Figure 4: Uninduced and induced SUMO-Peptide (signiferin BBa_K1022102)
CL: Cell lysate, M: Membrane pellet

Figure 5: SUMO-Peptide (signiferin BBa_K1022102) with IPTG induction
CL: Cell lysate, M: Membrane pellet

Figure 6: SUMO-Peptide (signiferin BBa_K1022102) solubilty
Lane 1: Low molecular weight marker, Lane 2: IPTG induced SUMO-peptide cell lysate fraction (CL)
Lane 4: Uninduced peptide-his without SUMO in cell lysate (CL), Lane 6:Uninduced peptide-his without SUMO in membrane pellet(M)
Lane 7:Induced peptide-his without SUMO in cell lysate (CL), Lane 9:Induced peptide-his without SUMO in membrane pellet (M)
Discussion
The SUMO domain was added to the peptide to improve its solubility [3]. This was really evident from the figure (4, 5). The peptides without SUMOylation did not have any over-expression around ~1.5 kDa (only peptide) in the cell lysate which is clearly seen in the figure 6 [ lanes 4,6 (uninduced) 7,9 (induced)] . But when a SUMO fusion of the same peptide was expressed they appeared as a thick band in the cell lysate as in the figure 6 [lane 2,black box] . From the figures [4,5] it is evident that there was some fraction of protein left with the cell debris. This clearly explains that SUMO does not increase the solubility to 100 % but just enhances the fraction of the soluble protein, thus facilitating easy purification. The band which was subjected to the MS/MS measurements clearly gave a 95 % sequence coverage to SUMO-peptide (signiferin) as in figure 7. Thus, we prove that the SUMO increases the solubility and also they are not toxic to the cell.

Figure 7: Sequence coverage from Tandem MS [Red letters]
Highlights
The SUMO domain that was used in our project was synthesized as a gene from DNA 2.0. The SUMO was designed to be bio-brick compatible without the restriction sites that are present in the prefix and suffix. The restriction sites where removed without disturbing the codons.
The Biobrick was designed in such a way that the ease of cloning SUMO fusion protein to be expressed has been improved. An Age I restriction site was introduced at the end of SUMO which when translated to amino acids Tyrosine Glycine Glycine. This is the recognition sequence of the Ulp-1 [4] protease that can cleave the peptides from the SUMO without any scar. So, one can use Age I and any one of the suffix enzyme to make a SUMO fusion.
SUMO cleavage by Ulp1
Cleavage of SUMO from Peptide by Ulp-1 protease.
The SUMO-peptide helps in increasing the soluble fraction of peptide but peptides are not biologically active in a fusion, they have to be cleaved from the fusion to get a active peptide fraction. This was achieved by in vivo production on SUMO specific Ulp-1 proteases.
A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter and the Ulp-1 production was driven by arabinose inducible promoter pBad. The plasmid was transformed into an BL21(DE3) pLysS strain. This construct was designed to check whether in vivo cleavage is possible. The main idea of the experiment is to first produce large amount of soluble fraction of SUMO-peptides and the produce the Ulp protease to cleave the sufficiently produced fusion peptides.

Figure 8: Part BBa_K1022116/117/118
Results
The presence of plasmid with gene inserts encoding the SUMO-peptide (BBa_K1022116, BBa_K1022117,BBa_K1022118) was confirmed by a colony pcr. The expected size of the insert was approximately around ~1500 bp which was clearly evident from the agarose gel picture figure(9,10,11). Though the actual size of the inserts are ~1300 bp the use of sequencing primers VF2 and VR which bind 100-150 base pair away from suffix and prefix the bands are around 1500 bp. An Eurogentec Smartladder MW-1700-10 [lane 1] (https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf) was used to identify the size of the fragments.

Figure 9: pBad Ulp SUMO maximin-H5 (BBa_K1022116) Colony PCR

Figure 10:pBad Ulp SUMO magainin (BBa_K1022118) colony PCR

Figure 11:pBad Ulp SUMO signiferin (BBa_K1022117) colony PCR
Discussion
The cleavage of the peptide from the SUMO is more crucial to free the peptide and make it biologically active. But if we look into the structure of the peptides, they are quite hydrophobic to be present as free peptides in the medium. So, a classical SDS page analysis is not suitable for these peptides. This made us to analyse our whole cell lysates with tandem MS approach. The cell lysates devoid of debris was subjected to MS/MS measurements. which gave a 40 % sequence coverage to the SUMO without the peptide as in figure 12. The free peptide was not present intact in the solution. This could be attributed to the hydrophobicity of the free peptide that makes it really difficult as they are not present free in the solution. But we could clearly prove that the Ulp-1 protease cleaved the peptide from the SUMO.

Figure 12:Coverage for SUMO without Peptide from Tandem MS [Red Letters]
Future Insights
Some intense literature studies gave us some insights of how we can improve the design of the peptides. Usually, the frogs and toads have the peptides with some post translational modifications (PTM) like amidation [5],polyglycylation [6] ,etc. These make them less vulnerable to carboxypeptidases or amidopeptidases. But, the major hurdle is that the E. coli cannot perform these PTMs that made us choose peptides without PTMs. This makes it clear that there are more possibilities that the peptides could be cleave of by the proteases.
We also found that some of the antimicrobial peptide (AMP) group called bombinins [7] was similar with maximins, which by itself is from the Bombina species. But, the bombinins have D-isoleucine instead of L-isoleucine. This makes it difficult for the peptidases to cleave the peptides. So, the maximins could also have some D-aminoacids that makes it much more difficult to perform these PTMs using E. coli.
Thus, in near future these peptides can be made if we can manage to make create a strain that can perform these PTMs so we can use them as a chassis to produce our peptides
References
- Michael R. Yeaman & Nannette Y. Yount,..(2003) 'Mechanisms of Antimicrobial Peptide Action and Resistance' , Pharmacological Reviews,vol 55,p:27-55
- Wang, Z. and Wang, G.* (2004) APD: the Antimicrobial Peptide Database. Nucleic Acids Research 32, D590-D592
- Panavas T, Sanders C, Butt TR.. (2009). SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods in Molecular Biology. 10 (8), p4-20.
- Lee, C., Sun, H., Hu, S., Chiu, C., Homhuan, A., Liang, S., Leng, C. and Wang, T. 2008. An improved SUMO fusion protein system for effective production of native proteins. Protein Science, 17 (7), pp. 1241--1248.
- Li, Recombinant production of antimicrobial peptides in Escherichia coli: A review. Protein Expression and Purification, 80, 2, 260-267, 2011.
- Redeker, V. 2010. Mass spectrometry analysis of C-terminal posttranslational modifications of tubulins.Methods in cell biology, 95 pp. 77--103.
- Mangoni, M., Grovale, N., Giorgi, A., Mignogna, G., Simmaco, M. and Barra, D. 2000. Structure-function relationships in bombinins H, antimicrobial peptides from Bombina skin secretions.Peptides, 21 (11), pp. 1673--1679.
 "
"
