Team:TU-Munich/Results/Moss
From 2013.igem.org
LouiseFunke (Talk | contribs) (→Growing Physcomitrella patens on solid materials) |
(→Tolerance to relevant environmental pollutants and toxins) |
||
| (18 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<!-- Start of content --> | <!-- Start of content --> | ||
| - | == | + | == Creation of transgenic ''Physcomitrella patens'' plants == |
| - | For implementation of the PhyscoFilter it is elementary to know about the mosses growth behavior on different surfaces. We therefore tried 5 different materials and came to the conclusion that the moss grows very well on all of them, but especially felt material would suit our plans for implementing the filter system in form of a [https://2013.igem.org/Team:TU-Munich/ | + | [[File:TUM13_Workflow_Transformation.png|thumb|center|910px|'''Figure 1:''' Work flow for the generation of transgenic moss.]] |
| + | |||
| + | ===1. Generation of expression constructs=== | ||
| + | |||
| + | {|cellspacing="0" border="1" right | ||
| + | |+ '''Table 1:''' Transgenic ''Physcomitrella patens'' plants | ||
| + | !Number | ||
| + | !Construct name (abbreviation) | ||
| + | !BioBrick | ||
| + | !Successful Transformation? | ||
| + | |- | ||
| + | |PF-1 | ||
| + | |GFP (GFP-cyt) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-2 | ||
| + | |Igk-GFP (GFP-sec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159304 BBa_K1159304] + [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-3 | ||
| + | |NanoLuciferase (nLuc-cyt) | ||
| + | |<partinfo>BBa_K1159001</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-4 | ||
| + | |Igk-NanoLuciferase (nLuc-sec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159006 BBa_K1159006] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-5 | ||
| + | |SERK-NanoLuciferase (SERK-nLuc) | ||
| + | |<partinfo>BBa_K1159010</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-6 | ||
| + | |NanoLuciferase-Receptor (nLuc-rec) | ||
| + | |<partinfo>BBa_K1159015</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-7 | ||
| + | |Erythromycinesterase (EreB-cyt) | ||
| + | |<partinfo>BBa_K1159000</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-8 | ||
| + | |Erythromycinesterase-Receptor (EreB-rec) | ||
| + | |<partinfo>BBa_K1159014</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-9 | ||
| + | |Ig Kappa Erythromycinesterase (EreB-sec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159005 BBa_K1159005] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-10 | ||
| + | |Laccase-Receptor (Lac-rec) | ||
| + | |<partinfo>BBa_K1159016</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-11 | ||
| + | |Ig Kappa Laccase (Lac-sec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159007 BBa_K1159007] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-12 | ||
| + | |Catecholdioxygenase (XylE-cyt) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159012 BBa_K1159012] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-13 | ||
| + | |DDT-dehydrochlorinase (GST-cyt) | ||
| + | |<partinfo>BBa_E0040</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-14 | ||
| + | |PP1-Receptor (PP1-rec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159019 BBa_K1159019] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-15 | ||
| + | |FluA-Receptor (FluA-rec) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159017 BBa_K1159017] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-16 | ||
| + | |Stressinducible_Promoter-RFP (Stress) | ||
| + | |<partinfo>BBa_E0040</partinfo> | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-17 | ||
| + | |SpyCatcher-Receptor:Spytag-nLuc (Catcher:Tag-nLuc) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159212 BBa_K1159212] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-18 | ||
| + | |SpyCatcher-Receptor:nLuc-Spytag (Catcher:nLuc-Tag) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159213 BBa_K1159213] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-19 | ||
| + | |SpyTag-Receptor:SpyCatcher-nLuc (Tag:Catcher-nLuc) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159214 BBa_K1159214] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-20 | ||
| + | |SpyTag-Receptor:nLuc-SpyCatcher (Tag:nLuc-Catcher) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159215 BBa_K1159215] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-21 | ||
| + | |Alcohol acetyltransferase I (Banana) | ||
| + | |[http://parts.igem.org/Part:BBa_J45014 BBa_J45014] | ||
| + | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html> | ||
| + | |- | ||
| + | |PF-22 | ||
| + | |Kill-switch with PIF3 (PIF-3) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159107 BBa_K1159107] | ||
| + | |no | ||
| + | |- | ||
| + | |PF-23 | ||
| + | |Kill-switch with PIF6 (PIF-6) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159108 BBa_K1159108] | ||
| + | | no | ||
| + | |- | ||
| + | |PF-24 | ||
| + | |Kill-switch with PIF6 (FRET) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159120 BBa_K1159120] | ||
| + | | no | ||
| + | |- | ||
| + | |PF-25 | ||
| + | |Kill-switch with PIF3 (FRET) | ||
| + | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159119 BBa_K1159119] | ||
| + | | no | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ===2. Preparation of linear DNA=== | ||
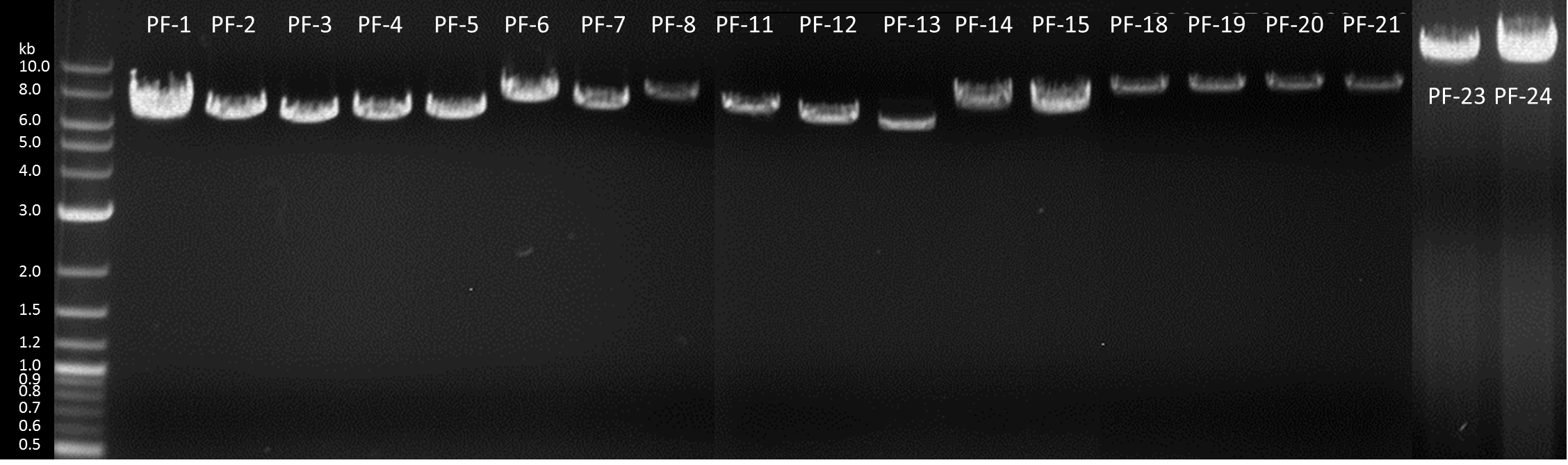
| + | In order to transfrom the moss, the DNA including our BioBricks with the nptII selection casette had to be linearized. We used EcoRI for the preparative digestion of our midipreps and simultaneously ran an analytical digest as quality control. Our preparations continued by purifying our digested DNA through salt, isopropanol and ethanol precipitation, solubilising in a small amount of water, measuring the concentration at the NanoDrop and suspending it in different volumes of sterile filtered Ca(NO<sub>3</sub>)<sub>2</sub> buffer for the intended concentration of 0.25 µg/µl . | ||
| + | [[File:TUM13_Midiprep_picture1.png|thumb|left|450px|'''Figure 2''': Louise and Christopher preparing midipreps]] | ||
| + | [[File:TUM13 Midiprep picture2.png|thumb|right|420px|'''Figure 3''': Midipreps for the moss transformation]] | ||
| + | [[File:TUM13_Midiprepgel.png|thumb|center|910px|'''Figure 4''': Analytical gelelectrophoresis of linearized DNA constructs]] | ||
| + | |||
| + | ===3. Transformation of ''Physcomitrella patens''=== | ||
| + | To transform Physcomitrella patens, the moss material has to be taken from the liquid culture and its cell walls have to be digested with driselase dissolved in mannitol to obtain protoplasts. The protoplasts are isolated by passing the digested material through sieves and the enzyme is washed off with mannitol and then resuspended in mannitol. | ||
| + | |||
| + | The number of protoplasts is determined with a hemocytometer and the material is suspended in the right amount of 3M medium to adjust the concentration. The linearized and purified DNA is mixed with PEG4000 and the protoplast solution and incubated while regularly mixing. After incubation, the mixture is diluted with 3M medium, centrifuged and resuspended in regeneration medium. | ||
| + | |||
| + | The protoplasts are put into 6-well plates, left in the dark over night and then left for 10 days for the regeneration of the cell walls at standard conditions. After moving the protoplasts onto solid medium covered with a layer of cellophane for three days, they are transferred to solid selection medium plates for two weeks. To ensure stabile integration, repeat the two weeks of selection after a two week release phase. | ||
| + | |||
| + | ====Trips to Freiburg ==== | ||
| + | <html> | ||
| + | <div class="box-center"> | ||
| + | <ul class="bxgallery"> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/48/TUM13_Foto_Freiburgvisit1.jpg/350px-TUM13_Foto_Freiburgvisit1.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/41/TUM13_Foto_Freiburgvisit2.jpg/350px-TUM13_Foto_Freiburgvisit2.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/d8/TUM13_Foto_Freiburgvisit3.jpg/350px-TUM13_Foto_Freiburgvisit3.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/25/TUM13_Foto_Freiburgvisit4.jpg/350px-TUM13_Foto_Freiburgvisit4.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/13/TUM13_Foto_Freiburgvisit5.jpg/350px-TUM13_Foto_Freiburgvisit5.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/50/TUM13_Foto_Freiburgvisit6.jpg/350px-TUM13_Foto_Freiburgvisit6.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/27/TUM13_Foto_Freiburgvisit7.jpg/350px-TUM13_Foto_Freiburgvisit7.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/46/TUM13_Foto_Freiburgvisit8.jpg/350px-TUM13_Foto_Freiburgvisit8.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/df/TUM13_Foto_Freiburgvisit9.jpg/350px-TUM13_Foto_Freiburgvisit9.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/2e/TUM13_Foto_Freiburgvisit10.jpg/350px-TUM13_Foto_Freiburgvisit10.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/26/TUM13_Foto_Freiburgvisit11.jpg/350px-TUM13_Foto_Freiburgvisit11.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/de/TUM13_Foto_Freiburgvisit12.jpg/350px-TUM13_Foto_Freiburgvisit12.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3b/TUM13_Foto_Freiburgvisit13.jpg/350px-TUM13_Foto_Freiburgvisit13.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/e/ec/TUM13_Foto_Freiburgvisit14.jpg/350px-TUM13_Foto_Freiburgvisit14.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/e/e6/TUM13_Foto_Freiburgvisit15.jpg/350px-TUM13_Foto_Freiburgvisit15.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/7/70/TUM13_Foto_Freiburgvisit16.jpg/350px-TUM13_Foto_Freiburgvisit16.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/8/8e/TUM13_Foto_Freiburgvisit17.jpg/350px-TUM13_Foto_Freiburgvisit17.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/b/b9/TUM13_Foto_Freiburgvisit18.jpg/350px-TUM13_Foto_Freiburgvisit18.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/94/TUM13_Foto_Freiburgvisit19.jpg/350px-TUM13_Foto_Freiburgvisit19.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/fc/TUM13_Foto_Freiburgvisit20.jpg/350px-TUM13_Foto_Freiburgvisit20.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/54/TUM13_Foto_Freiburgvisit21.jpg/350px-TUM13_Foto_Freiburgvisit21.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/5a/TUM13_Foto_Freiburgvisit22.jpg/350px-TUM13_Foto_Freiburgvisit22.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/23/TUM13_Foto_Freiburgvisit23.jpg/350px-TUM13_Foto_Freiburgvisit23.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/91/TUM13_Foto_Freiburgvisit24.jpg/350px-TUM13_Foto_Freiburgvisit24.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/8/81/TUM13_Foto_Freiburgvisit25.jpg/350px-TUM13_Foto_Freiburgvisit25.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/f9/TUM13_Foto_Freiburgvisit26.jpg/350px-TUM13_Foto_Freiburgvisit26.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3b/TUM13_Foto_Freiburgvisit27.jpg/350px-TUM13_Foto_Freiburgvisit27.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/f2/TUM13_Foto_Freiburgvisit28.jpg/350px-TUM13_Foto_Freiburgvisit28.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/0/0d/TUM13_Foto_Freiburgvisit29.jpg/350px-TUM13_Foto_Freiburgvisit29.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/a7/TUM13_Foto_Freiburgvisit30.jpg/350px-TUM13_Foto_Freiburgvisit30.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/ab/TUM13_Foto_Freiburgvisit31.jpg/350px-TUM13_Foto_Freiburgvisit31.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3d/TUM13_Foto_Freiburgvisit32.jpg/350px-TUM13_Foto_Freiburgvisit32.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/0/06/TUM13_Foto_Freiburgvisit33.jpg/350px-TUM13_Foto_Freiburgvisit33.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/12/TUM13_Foto_Freiburgvisit34.jpg/350px-TUM13_Foto_Freiburgvisit34.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/31/TUM13_Foto_Freiburgvisit35.jpg/350px-TUM13_Foto_Freiburgvisit35.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/16/TUM13_Foto_Freiburgvisit36.jpg/350px-TUM13_Foto_Freiburgvisit36.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/95/TUM13_Foto_Freiburgvisit37.jpg/350px-TUM13_Foto_Freiburgvisit37.jpg" /></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/ac/TUM13_Foto_Freiburgvisit38.jpg/350px-TUM13_Foto_Freiburgvisit38.jpg" /></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | We had the great chance to perform our '' '''Physcomitrella patens''' '' transformations at Prof. Dr.Reski´s lab in Freiburg, with Dr. Wiedemann as our expert instructor. Our first trip started in a great hurry, because we worked on our DNA preparations until the very last minute. Ingmar even pulled an all-nighter to get the DNA ready. We would have missed our intercity bus if it wasn’t for Rosario who drove us to the bus station, all squished together in the “pizza mobile” with the trunk full with our medium bottles and lab equipment. After five hours on the bus, we fell into bed to get some rest, because we had a very long day ahead of us. | ||
| + | |||
| + | We arrived at the Reski lab early in the morning to meet with Dr. Gertrud Wiedemann, who instructed us throughout the day and gave us many tips how to proceed with the moss. Because of the incubation times and because it was our first try, it took us ten hours without a break until we had two boxes stacked with 6 well plates. We quickly went to get some beers and chips and met with the Freiburg iGEM team for a really nice barbecue. When we finally left, public transport wasn’t running anymore, so we didn’t miss the chance to take a midnight sightseeing tour through Freiburg, where Volker showed us around. | ||
| + | |||
| + | Ten days later, Johanna and Andi visited the Freiburg lab to transfer the then regenerated protoplasts onto agar plates and soon after, we came back for our second and final round of transformation. At our first try, we didn’t get enough moss protoplasts, so we worked through two batches and therefore had to prepare another batch of driselase. Our handling had improved, yet it still took twelve hours and again, there was no time left for a break. After we said good bye, we celebrated with a couple of beers and some yummy flammkuchen at the UC uni café of Freiburg. We had learned so much and got much closer to our goal. A big successful step for our team! | ||
| + | |||
| + | ===4. Regeneration and Selection of transgenic plants=== | ||
| + | After Transformation, the regenerating protoplasts were incubated in regeneration medium in 6-well-plates sealed with parafilm for 10 days and then cultivated on a layer of autoclaved cellophane (seperated with Whatman paper during autoclavation, so they dont stick together), on top of Knop medium agar plates for three days under standard conditions. Then we transferred the cellophane layers with the regenerated moss onto Knop medium agar plates containing 25 µg/ml G418 antibiotic for two weeks. The official protocol schedules a two week release period followed by another selection period to ensure stabile transormation, but we didn´t have that much time and went with a single round of selection. We plated only half of our transformed protoplasts. The other half was left in the 6-well-plates where 2 ml of selection medium were added to the 2 ml of regeneration medium from the transormation, with G418 diluted 1:8000. | ||
| + | |||
| + | == Testing cultivation conditions == | ||
| + | ===Growing ''Physcomitrella patens'' on solid materials=== | ||
| + | |||
| + | For implementation of the PhyscoFilter it is elementary to know about the mosses growth behavior on different surfaces. We therefore tried 5 different materials and came to the conclusion that the moss grows very well on all of them, but especially felt material would suit our plans for implementing the filter system in form of a [https://2013.igem.org/Team:TU-Munich/Results/Implementation#Our_swimming_remediation_raft remediation raft] since the moss can easily cling to the fibers, which are also dense enough to prevent the moss from being washed away. Also the spongy properties of felt make it an ideal surface for the plant as it ensures a constant supply of water. The second best material is probably agar or metal grid on agar, but this has the disadvantage of being washed away gradually. | ||
<!--- | <!--- | ||
[[File:TUM13_surface1.png|thumb|350px]] | [[File:TUM13_surface1.png|thumb|350px]] | ||
| Line 33: | Line 242: | ||
</html> | </html> | ||
| - | ==Determination of growth rates for different liquid culture forms== | + | ===Determination of growth rates for different liquid culture forms=== |
In parallel to the determination of growth conditions for solid materials we examined different growth conditions in liquid culture, too. In general the handling of liquid cultures of moss is more easy than the one of those growing on solid media as the moss can be disrupted mechanically with an Ultra-Turrax. That way homogenized cultures can be achieved very comfortably. Moreover the growth of moss in liquid suspension cultures provides an improved and constant nourishment of cells without any nutrition gradients and continuously pH adjusted. Even if the up scaling process up to volumes of around 20 L in standard stirred tank bioreactors is more convenient at a first glance, the further up scaling process is often limited by an insufficient light input resulting in suboptimal growth rates. This is physically determined as the volume increases in the third potency whereas the surface only increases in the second potency. Therefore larger suspension volumes require different and often technically more challenging bioreactor forms as tube reactors, plate or wave reactors. | In parallel to the determination of growth conditions for solid materials we examined different growth conditions in liquid culture, too. In general the handling of liquid cultures of moss is more easy than the one of those growing on solid media as the moss can be disrupted mechanically with an Ultra-Turrax. That way homogenized cultures can be achieved very comfortably. Moreover the growth of moss in liquid suspension cultures provides an improved and constant nourishment of cells without any nutrition gradients and continuously pH adjusted. Even if the up scaling process up to volumes of around 20 L in standard stirred tank bioreactors is more convenient at a first glance, the further up scaling process is often limited by an insufficient light input resulting in suboptimal growth rates. This is physically determined as the volume increases in the third potency whereas the surface only increases in the second potency. Therefore larger suspension volumes require different and often technically more challenging bioreactor forms as tube reactors, plate or wave reactors. | ||
<html> | <html> | ||
| Line 48: | Line 257: | ||
In our approaches we tested mainly the influence of mixing and coupled to this of aeration on the growth of moss. For this purpose 500 mL flasks containing 250 mL Knob media were inoculated with 50 mL moss suspension which had been disrupted 24 h ago (corresponding a moss concentration of 80 mg dry mass per liter). In triplicates the growth conditions in standing, shaken and aerated flasks were determined for 9 days at room temperature and a normal dark/light rhythm (8/16h). | In our approaches we tested mainly the influence of mixing and coupled to this of aeration on the growth of moss. For this purpose 500 mL flasks containing 250 mL Knob media were inoculated with 50 mL moss suspension which had been disrupted 24 h ago (corresponding a moss concentration of 80 mg dry mass per liter). In triplicates the growth conditions in standing, shaken and aerated flasks were determined for 9 days at room temperature and a normal dark/light rhythm (8/16h). | ||
| - | [[File:TUM_13__growth_conditions.png|thumb|left|340px| '''Figure | + | [[File:TUM_13__growth_conditions.png|thumb|left|340px| '''Figure 5''': Different experimental setups for the growth of wild type moss in liquid culture. '''A:''' Aerated flasks, '''B:''' Standing flask, '''C:''' Shaken flask]] |
| - | [[File:TUM13_growth_liquid_culture.png|thumb|right|520px| '''Figure | + | [[File:TUM13_growth_liquid_culture.png|thumb|right|520px| '''Figure 6:''' Dryweight determined for the different cultivation methods after nine days of incubation at normal growth conditions.]] |
All flasks were inoculated with 80 mg dry weight per litre moss. After nine days of incubation the biomass in the standing flask stayed approximately constant (82 mg per litre) compared to the beginning of the experiment. The biomasses in the shaken and in the aerated flasks increased in the same time to 118 and 168 mg per litre respectively. As all flaks were incubated under the same temperature and illumination conditions the internal mixing and especially linked to this the aeration seems to be of importance for biomass generation. | All flasks were inoculated with 80 mg dry weight per litre moss. After nine days of incubation the biomass in the standing flask stayed approximately constant (82 mg per litre) compared to the beginning of the experiment. The biomasses in the shaken and in the aerated flasks increased in the same time to 118 and 168 mg per litre respectively. As all flaks were incubated under the same temperature and illumination conditions the internal mixing and especially linked to this the aeration seems to be of importance for biomass generation. | ||
Normally at the [https://static.igem.org/mediawiki/2013/1/18/Physcomitrella_cell_culture_conditions.pdf Reski laboratory] the bioreactors are aerated with 0,3 vvm at a light intensity of 55 μmol m-2 s-1 . So far unpublished results indicate that aeration with up to 6 volume percent carbon dioxide improves the growth rate if the light intensity is increased as well. Nevertheless an increased light intensity automatically requires a stronger cooling capacity of the bioreactor due to photons which are not absorbed by the photo systems. Therefore the development of illumination which only serves the wavelengths required by the photosystems in plants could be an interesting alternative. Moreover the addition of further carbon sources than carbon dioxide could boost the growth of the moss as well. So far the addition of glucose leads to a change of colour to brown of the moss plants if applied for longer than 14 days. An optimization of the media composition as well as the testing of different feeding strategies could help to solve this problem. | Normally at the [https://static.igem.org/mediawiki/2013/1/18/Physcomitrella_cell_culture_conditions.pdf Reski laboratory] the bioreactors are aerated with 0,3 vvm at a light intensity of 55 μmol m-2 s-1 . So far unpublished results indicate that aeration with up to 6 volume percent carbon dioxide improves the growth rate if the light intensity is increased as well. Nevertheless an increased light intensity automatically requires a stronger cooling capacity of the bioreactor due to photons which are not absorbed by the photo systems. Therefore the development of illumination which only serves the wavelengths required by the photosystems in plants could be an interesting alternative. Moreover the addition of further carbon sources than carbon dioxide could boost the growth of the moss as well. So far the addition of glucose leads to a change of colour to brown of the moss plants if applied for longer than 14 days. An optimization of the media composition as well as the testing of different feeding strategies could help to solve this problem. | ||
| - | ==Tolerance to relevant environmental pollutants and toxins== | + | ===Tolerance to relevant environmental pollutants and toxins=== |
To test whether and how the moss reacts to toxins and pollutants, which can occur in waste and surface water and which our PhyscoFilter should remove, wild type plants were incubated in serial dilutions of the toxic substances. As a negative control distilled water was used. After 4, 7, 10 and 19 days the plants were screened with a light microscope, where one could easily differentiate between alive and dead plants. The latter occurred in two different phenotypes, one appearing transparent (dead moss 1) because it lost its chlorophyll, the other black (dead moss 2). | To test whether and how the moss reacts to toxins and pollutants, which can occur in waste and surface water and which our PhyscoFilter should remove, wild type plants were incubated in serial dilutions of the toxic substances. As a negative control distilled water was used. After 4, 7, 10 and 19 days the plants were screened with a light microscope, where one could easily differentiate between alive and dead plants. The latter occurred in two different phenotypes, one appearing transparent (dead moss 1) because it lost its chlorophyll, the other black (dead moss 2). | ||
| - | [[File:TUM13_toxassay.png|thumb|center|900px| '''Figure | + | [[File:TUM13_toxassay.png|thumb|center|900px| '''Figure 7:''' Toxicity assay for wild type moss]] |
<div class="visualClear"></div> | <div class="visualClear"></div> | ||
{| | {| | ||
| - | |+ Table | + | |+ '''Table 2''': Typical concentrations of toxic substances |
! Substance | ! Substance | ||
! Application | ! Application | ||
| Line 110: | Line 319: | ||
==References:== | ==References:== | ||
| - | [[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf University of Toledo, Department of Environmental Sciences]] | + | [[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf University of Toledo, Department of Environmental Sciences]] Working concentrations of various antibiotics |
| - | [[http://www.plant-biotech.net plant-biotech.net ]] | + | |
| - | [[https://static.igem.org/mediawiki/2013/1/18/Physcomitrella_cell_culture_conditions.pdf Physcomitrella cell culture conditions]] | + | [[http://www.plant-biotech.net plant-biotech.net ]] Prof. Reski´s Homepage |
| - | [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586047/ Liao,Y. et al, 2006]] Liao,Y. et al, (2006). The Key Role of Chlorocatechol 1,2-Dioxygenase in Phytoremoval and Degradation of Catechol by Transgenic Arabidopsis. ''Plant Physiology'', 142(2): 620–628 | + | |
| + | [[https://static.igem.org/mediawiki/2013/1/18/Physcomitrella_cell_culture_conditions.pdf Physcomitrella cell culture conditions]] | ||
| + | |||
| + | [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586047/ Liao,Y. et al, 2006]] Liao,Y. et al, (2006). The Key Role of Chlorocatechol 1,2-Dioxygenase in Phytoremoval and Degradation of Catechol by Transgenic Arabidopsis. ''Plant Physiology'', 142(2): 620–628 | ||
<!-- End of content --> | <!-- End of content --> | ||
Latest revision as of 03:23, 29 October 2013
Creation of transgenic Physcomitrella patens plants
1. Generation of expression constructs
| Number | Construct name (abbreviation) | BioBrick | Successful Transformation? |
|---|---|---|---|
| PF-1 | GFP (GFP-cyt) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311] | 
|
| PF-2 | Igk-GFP (GFP-sec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159304 BBa_K1159304] + [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311] | 
|
| PF-3 | NanoLuciferase (nLuc-cyt) | <partinfo>BBa_K1159001</partinfo> | 
|
| PF-4 | Igk-NanoLuciferase (nLuc-sec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159006 BBa_K1159006] | 
|
| PF-5 | SERK-NanoLuciferase (SERK-nLuc) | <partinfo>BBa_K1159010</partinfo> | 
|
| PF-6 | NanoLuciferase-Receptor (nLuc-rec) | <partinfo>BBa_K1159015</partinfo> | 
|
| PF-7 | Erythromycinesterase (EreB-cyt) | <partinfo>BBa_K1159000</partinfo> | 
|
| PF-8 | Erythromycinesterase-Receptor (EreB-rec) | <partinfo>BBa_K1159014</partinfo> | 
|
| PF-9 | Ig Kappa Erythromycinesterase (EreB-sec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159005 BBa_K1159005] | 
|
| PF-10 | Laccase-Receptor (Lac-rec) | <partinfo>BBa_K1159016</partinfo> | 
|
| PF-11 | Ig Kappa Laccase (Lac-sec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159007 BBa_K1159007] | 
|
| PF-12 | Catecholdioxygenase (XylE-cyt) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159012 BBa_K1159012] | 
|
| PF-13 | DDT-dehydrochlorinase (GST-cyt) | <partinfo>BBa_E0040</partinfo> | 
|
| PF-14 | PP1-Receptor (PP1-rec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159019 BBa_K1159019] | 
|
| PF-15 | FluA-Receptor (FluA-rec) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159017 BBa_K1159017] | 
|
| PF-16 | Stressinducible_Promoter-RFP (Stress) | <partinfo>BBa_E0040</partinfo> | 
|
| PF-17 | SpyCatcher-Receptor:Spytag-nLuc (Catcher:Tag-nLuc) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159212 BBa_K1159212] | 
|
| PF-18 | SpyCatcher-Receptor:nLuc-Spytag (Catcher:nLuc-Tag) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159213 BBa_K1159213] | 
|
| PF-19 | SpyTag-Receptor:SpyCatcher-nLuc (Tag:Catcher-nLuc) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159214 BBa_K1159214] | 
|
| PF-20 | SpyTag-Receptor:nLuc-SpyCatcher (Tag:nLuc-Catcher) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159215 BBa_K1159215] | 
|
| PF-21 | Alcohol acetyltransferase I (Banana) | [http://parts.igem.org/Part:BBa_J45014 BBa_J45014] | 
|
| PF-22 | Kill-switch with PIF3 (PIF-3) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159107 BBa_K1159107] | no |
| PF-23 | Kill-switch with PIF6 (PIF-6) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159108 BBa_K1159108] | no |
| PF-24 | Kill-switch with PIF6 (FRET) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159120 BBa_K1159120] | no |
| PF-25 | Kill-switch with PIF3 (FRET) | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159119 BBa_K1159119] | no |
2. Preparation of linear DNA
In order to transfrom the moss, the DNA including our BioBricks with the nptII selection casette had to be linearized. We used EcoRI for the preparative digestion of our midipreps and simultaneously ran an analytical digest as quality control. Our preparations continued by purifying our digested DNA through salt, isopropanol and ethanol precipitation, solubilising in a small amount of water, measuring the concentration at the NanoDrop and suspending it in different volumes of sterile filtered Ca(NO3)2 buffer for the intended concentration of 0.25 µg/µl .
3. Transformation of Physcomitrella patens
To transform Physcomitrella patens, the moss material has to be taken from the liquid culture and its cell walls have to be digested with driselase dissolved in mannitol to obtain protoplasts. The protoplasts are isolated by passing the digested material through sieves and the enzyme is washed off with mannitol and then resuspended in mannitol.
The number of protoplasts is determined with a hemocytometer and the material is suspended in the right amount of 3M medium to adjust the concentration. The linearized and purified DNA is mixed with PEG4000 and the protoplast solution and incubated while regularly mixing. After incubation, the mixture is diluted with 3M medium, centrifuged and resuspended in regeneration medium.
The protoplasts are put into 6-well plates, left in the dark over night and then left for 10 days for the regeneration of the cell walls at standard conditions. After moving the protoplasts onto solid medium covered with a layer of cellophane for three days, they are transferred to solid selection medium plates for two weeks. To ensure stabile integration, repeat the two weeks of selection after a two week release phase.
Trips to Freiburg
We had the great chance to perform our Physcomitrella patens transformations at Prof. Dr.Reski´s lab in Freiburg, with Dr. Wiedemann as our expert instructor. Our first trip started in a great hurry, because we worked on our DNA preparations until the very last minute. Ingmar even pulled an all-nighter to get the DNA ready. We would have missed our intercity bus if it wasn’t for Rosario who drove us to the bus station, all squished together in the “pizza mobile” with the trunk full with our medium bottles and lab equipment. After five hours on the bus, we fell into bed to get some rest, because we had a very long day ahead of us.
We arrived at the Reski lab early in the morning to meet with Dr. Gertrud Wiedemann, who instructed us throughout the day and gave us many tips how to proceed with the moss. Because of the incubation times and because it was our first try, it took us ten hours without a break until we had two boxes stacked with 6 well plates. We quickly went to get some beers and chips and met with the Freiburg iGEM team for a really nice barbecue. When we finally left, public transport wasn’t running anymore, so we didn’t miss the chance to take a midnight sightseeing tour through Freiburg, where Volker showed us around.
Ten days later, Johanna and Andi visited the Freiburg lab to transfer the then regenerated protoplasts onto agar plates and soon after, we came back for our second and final round of transformation. At our first try, we didn’t get enough moss protoplasts, so we worked through two batches and therefore had to prepare another batch of driselase. Our handling had improved, yet it still took twelve hours and again, there was no time left for a break. After we said good bye, we celebrated with a couple of beers and some yummy flammkuchen at the UC uni café of Freiburg. We had learned so much and got much closer to our goal. A big successful step for our team!
4. Regeneration and Selection of transgenic plants
After Transformation, the regenerating protoplasts were incubated in regeneration medium in 6-well-plates sealed with parafilm for 10 days and then cultivated on a layer of autoclaved cellophane (seperated with Whatman paper during autoclavation, so they dont stick together), on top of Knop medium agar plates for three days under standard conditions. Then we transferred the cellophane layers with the regenerated moss onto Knop medium agar plates containing 25 µg/ml G418 antibiotic for two weeks. The official protocol schedules a two week release period followed by another selection period to ensure stabile transormation, but we didn´t have that much time and went with a single round of selection. We plated only half of our transformed protoplasts. The other half was left in the 6-well-plates where 2 ml of selection medium were added to the 2 ml of regeneration medium from the transormation, with G418 diluted 1:8000.
Testing cultivation conditions
Growing Physcomitrella patens on solid materials
For implementation of the PhyscoFilter it is elementary to know about the mosses growth behavior on different surfaces. We therefore tried 5 different materials and came to the conclusion that the moss grows very well on all of them, but especially felt material would suit our plans for implementing the filter system in form of a remediation raft since the moss can easily cling to the fibers, which are also dense enough to prevent the moss from being washed away. Also the spongy properties of felt make it an ideal surface for the plant as it ensures a constant supply of water. The second best material is probably agar or metal grid on agar, but this has the disadvantage of being washed away gradually.
Determination of growth rates for different liquid culture forms
In parallel to the determination of growth conditions for solid materials we examined different growth conditions in liquid culture, too. In general the handling of liquid cultures of moss is more easy than the one of those growing on solid media as the moss can be disrupted mechanically with an Ultra-Turrax. That way homogenized cultures can be achieved very comfortably. Moreover the growth of moss in liquid suspension cultures provides an improved and constant nourishment of cells without any nutrition gradients and continuously pH adjusted. Even if the up scaling process up to volumes of around 20 L in standard stirred tank bioreactors is more convenient at a first glance, the further up scaling process is often limited by an insufficient light input resulting in suboptimal growth rates. This is physically determined as the volume increases in the third potency whereas the surface only increases in the second potency. Therefore larger suspension volumes require different and often technically more challenging bioreactor forms as tube reactors, plate or wave reactors.
In our approaches we tested mainly the influence of mixing and coupled to this of aeration on the growth of moss. For this purpose 500 mL flasks containing 250 mL Knob media were inoculated with 50 mL moss suspension which had been disrupted 24 h ago (corresponding a moss concentration of 80 mg dry mass per liter). In triplicates the growth conditions in standing, shaken and aerated flasks were determined for 9 days at room temperature and a normal dark/light rhythm (8/16h).
All flasks were inoculated with 80 mg dry weight per litre moss. After nine days of incubation the biomass in the standing flask stayed approximately constant (82 mg per litre) compared to the beginning of the experiment. The biomasses in the shaken and in the aerated flasks increased in the same time to 118 and 168 mg per litre respectively. As all flaks were incubated under the same temperature and illumination conditions the internal mixing and especially linked to this the aeration seems to be of importance for biomass generation. Normally at the Reski laboratory the bioreactors are aerated with 0,3 vvm at a light intensity of 55 μmol m-2 s-1 . So far unpublished results indicate that aeration with up to 6 volume percent carbon dioxide improves the growth rate if the light intensity is increased as well. Nevertheless an increased light intensity automatically requires a stronger cooling capacity of the bioreactor due to photons which are not absorbed by the photo systems. Therefore the development of illumination which only serves the wavelengths required by the photosystems in plants could be an interesting alternative. Moreover the addition of further carbon sources than carbon dioxide could boost the growth of the moss as well. So far the addition of glucose leads to a change of colour to brown of the moss plants if applied for longer than 14 days. An optimization of the media composition as well as the testing of different feeding strategies could help to solve this problem.
Tolerance to relevant environmental pollutants and toxins
To test whether and how the moss reacts to toxins and pollutants, which can occur in waste and surface water and which our PhyscoFilter should remove, wild type plants were incubated in serial dilutions of the toxic substances. As a negative control distilled water was used. After 4, 7, 10 and 19 days the plants were screened with a light microscope, where one could easily differentiate between alive and dead plants. The latter occurred in two different phenotypes, one appearing transparent (dead moss 1) because it lost its chlorophyll, the other black (dead moss 2).
| Substance | Application | Concentration |
|---|---|---|
| Ampicillin | Antibiotic agar plate | 0.1 g/L[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf[1]] |
| Chloramphenicol | Antibiotic agar plate | 0.025 g/L[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf[1]] |
| Kanamycin | Antibiotic agar plate | 0.05 g/L[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf[1]] |
| Tetracycline | Antibiotic agar plate | 0.01 g/L[http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working%20concentrations%20and%20stock%20solutions.pdf[1]] |
| Geneticin (G418) | Antibiotic agar plate (for moss selection) | 12.5 mg/L[http://www.plant-biotech.net[2]] |
| Diclofenac | 1 tablet (25-50 mg) dissolved in 6 L (blood circuit) | 4.2-8.3 mg/L |
| NaCl | Sea water | 3.5 g/L |
| Catechol | Death of Arabidopsis | 55 mg/L[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586047/[3]] |
| Erythromycin | 1 tablet (500 mg) dissolved in 6 L (blood circuit) | 0.08 g/L |
The result of this toxicity assay is, that wild type plants are not negatively affected by waste water treatment plant (WWTP) effluents, which were sampled from the local WWTPs Großlappen (waste water 1) and Garching (waste water 2). So the filter system could work effectively placed in the effluent stream of WWTPs or on surface water. However, sea water seems to influence the vitality of the moss, so the implementation of the PhyscoFilter in salt water is not recommended. Furthermore we can conclude that substances the genetically modified moss should degrade (Erythromycin, Catechol) or accumulate (Diclofenac) only affect the plant - if they do at all - at concentrations much higher than they occur naturally (see table 1). Also the assay indicates that it is possible to grow the plant on agar plates with often used antibiotics (Tetracycline, Ampicillin, Chloramphenicol, Kanamycin), since the working concentration has no influence on the moss. This can be very useful to prevent bacterial contamination of plates. As expected G418 shows toxic influence on wild type moss and can therefore be used as selection substance for transformed plants, though it takes a few days to take effect.
References:
http://www.eeescience.utoledo.edu/Faculty/Sigler/Von_Sigler/LEPR_Protocols_files/Working concentrations and stock solutions.pdf University of Toledo, Department of Environmental Sciences Working concentrations of various antibiotics
http://www.plant-biotech.net plant-biotech.net Prof. Reski´s Homepage
[Physcomitrella cell culture conditions]
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586047/ Liao,Y. et al, 2006 Liao,Y. et al, (2006). The Key Role of Chlorocatechol 1,2-Dioxygenase in Phytoremoval and Degradation of Catechol by Transgenic Arabidopsis. Plant Physiology, 142(2): 620–628
 "
"


























































AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351