Team:TU-Munich/Project/Physcomitrella
From 2013.igem.org
(→Advantages as a new chassis for iGEM) |
PSchneider (Talk | contribs) m (→Moss methods - working with Physcomitrella) |
||
| (38 intermediate revisions not shown) | |||
| Line 48: | Line 48: | ||
===Expertbox: Prof. Reski=== | ===Expertbox: Prof. Reski=== | ||
| - | [[File:TUM13_expert_Reski.png|thumb|450px|left| '''Figure 3:''' Expert interview with Professor Reski. ]] When we | + | [[File:TUM13_expert_Reski.png|thumb|450px|left| '''Figure 3:''' Expert interview with Professor Reski. ]] |
| + | When we chose Phytoremediation as our project for this year´s competition, it soon became clear that ''Physcomitrella patens'' is a great chassis that could bring iGEM closer to the real world applications we were looking for. Therefore we contacted Prof. Dr. Reski, who is a recognized expert on ''Physcomitrella patens'' and its biotechnological applications. | ||
| + | |||
| + | Prof. Dr. Reski liked our idea of introducing his ''Physco'' to iGEM from the very beginning and offered his help wherever we would need it. Members of our team traveled to his lab in Freiburg (350 km) five times in total [https://2013.igem.org/Team:TU-Munich/Results/Moss#3._Transformation_of_Physcomitrella_patens] to fetch plasmids known to be functional in ''Physcomitrealla patens'' or for discussions with him or his co-workers. We could win him over as an advisor for our team, which shows how iGEM brings together universities and scientific groups from many different places. | ||
===Advantages as a new chassis for iGEM=== | ===Advantages as a new chassis for iGEM=== | ||
| Line 61: | Line 64: | ||
[[File:TUM13 Bench.png|thumb|right|350px|'''Figure 4:''' Working with cultivated moss on our laboratory bench]] | [[File:TUM13 Bench.png|thumb|right|350px|'''Figure 4:''' Working with cultivated moss on our laboratory bench]] | ||
| - | *<b>Cell culture</b>: ''Physcomitrella patens'' plants can be cultivated either on solidified medium or in liquid medium. Liquid cultures can be kept in Erlenmeyer flasks under constant rotation and light exposure or in bioreactors for large scale production. By regularly (ideally weekly) disrupting the plants mechanically with | + | *<b>Cell culture</b>: ''Physcomitrella patens'' plants can be cultivated either on solidified medium or in liquid medium. Liquid cultures can be kept in Erlenmeyer flasks under constant rotation and light exposure or in bioreactors for large scale production. By regularly (ideally weekly) disrupting the plants mechanically with a mixer (e.g. Ultra-Turrax) a homogenized culture can be achieved. |
*<b>Storage</b>: Long-term storage of ''Physcomitrella'' strains can be achieved by cryo-preservation. This procedure ensures a maximum survival rate of the plant by preconditioning and controlled freezing. [[http://www.plant-biotech.net/paper/PlantBiol_2004_Schulte_pageproof.pdf Schulte and Reski, 2004]] | *<b>Storage</b>: Long-term storage of ''Physcomitrella'' strains can be achieved by cryo-preservation. This procedure ensures a maximum survival rate of the plant by preconditioning and controlled freezing. [[http://www.plant-biotech.net/paper/PlantBiol_2004_Schulte_pageproof.pdf Schulte and Reski, 2004]] | ||
| - | *<b>Targeted knockout</b>: ''P. patens'' is unique among plants in its high efficiency of gene targeting by homologous recombination. To knockout a specific gene a | + | *<b>Targeted knockout</b>: ''P. patens'' is unique among plants in its high efficiency of gene targeting by homologous recombination. To knockout a specific gene a disruption construct has to be generated. This consists of the gene to be silenced with a selection cassette (usually the nptII gene) inserted into its center by suitable restriction sites. The moss is then transformed with this DNA construct which is integrated into the genome by homologous recombination. [[http://www.ncbi.nlm.nih.gov/pubmed/14586556 Reski et al., 2004]] |
| - | *<b>Transformation</b>: Transformation of ''P. patens'' requires protoplasts of the plant, which can be obtained by cell wall digesting enzymes. The protoplasts can either be transformed by particle bombardment or via polyethylene glycol (PEG), which is the easiest and most commonly used method. The DNA constructs should be linearized for optimal transformation and contain a selection marker for subsequent screening. After transformation the protoplasts are cultivated in the dark for 12-16 h followed by | + | [[File:TUM13 Transformation.png|thumb|right|350px|'''Figure 5:''' Transformation procedure]] |
| - | *<b>Analysis of transformants</b>: First | + | *<b>Transformation</b>: Transformation of ''P. patens'' requires protoplasts of the plant, which can be obtained by cell wall digesting enzymes. The protoplasts can either be transformed by particle bombardment or via polyethylene glycol (PEG), which is the easiest and most commonly used method. The DNA constructs should be linearized for optimal transformation and contain a selection marker for subsequent screening. After transformation, the protoplasts are cultivated in the dark for 12-16 h followed by 9 days under normal growth conditions during which they regenerate their cell wall. Afterwards they are plated on solidified medium in normal petri dishes and first experiments can be executed. [[http://link.springer.com/article/10.1007/BF00260654 Schaefer et al., 1991]] |
| + | *<b>Analysis of transformants</b>: First, the transformed protoplasts are plated on solidified medium containing the antibiotic G418 (Geneticin), to which successfully transformed plants are resistant due to the selection marker nptII which encodes the enzyme neomycin phosphotransferase. Secondly, the transformants are analyzed by a PCR screen using primers derived the selection marker cassette´s sequences [[http://www.plant-biotech.net/paper/PlantMolecularBiologyReporter_2002_Schween.pdf Schween et al., 2002]]. | ||
| - | [[File:TUM13_timeline.png|thumb| | + | [[File:TUM13_timeline.png|thumb|center|910px| '''Figure 6:''' Comparing work flow for ''E. coli'', ''S. cerevisiae'' and ''P. patens'']] |
| - | + | In '''conclusion''', it naturally takes a bit more time to work with a plant in comparison to bacteria or yeast (see figure 6), but the advantages definitely outweigh the time factor and if teams start early enough, it is perfectly possible to work with ''P. patens'' as a chassis in an iGEM project! | |
| - | + | This is a brief summary of the most important techniques in working with ''P. patens''. For a full description of these methods please see [https://2013.igem.org/Team:TU-Munich/Notebook/Methods#Physcomitrella_Methods our methods page]. | |
| - | ==BioBricks for | + | ==BioBricks for protein expression in ''Physcomitrella''== |
| - | [[File:TUM13_plant biobricks.png|thumb|right|300px| '''Figure | + | [[File:TUM13_plant biobricks.png|thumb|right|300px| '''Figure 7:''' Our transformation vector ]] |
| - | To transform the moss we created an all-purpose backbone into which all our constructs could be integrated easily. Therefore we modified the | + | To transform the moss, we created an all-purpose backbone into which all our constructs could be integrated easily. Therefore we modified the common iGEM pSB1C3 plasmid with the following plant-specific components: |
| - | * '''pAct5 | + | * '''pAct5 (Actin 5 promoter from ''Physcomitrella patens''):''' for strong protein expression. |
| - | * '''miniMCS''' | + | * '''miniMCS:''' This miniature multiple cloning site contains restriction sites for the enzymes MfeI and SbfI which allows you to insert BioBrick between two other Biobricks (in our case: pAct5 and t35S_npt-cassette) as a result of the compability of MfeI with EcoRI and SbfI with PstI. |
| - | * '''t35S (35S terminator)''' | + | * '''t35S (35S terminator):''' This terminator originates from the 35S transcript of the cauliflower mosaic virus. |
| - | * '''nptII-cassette''' | + | * '''nptII-cassette:''' This selection cassette contains the neomycon phosphotransferase II gene, encoding the enzyme aminoglycoside-3'-phosphotransferase (NPTII), which inactivates a range of aminoglycoside antibiotics such as Kanamycin, Neomycin and Geneticin (G418) by phosphorylation. The latter antibiotic is used as selective agent for transformed ''P. patens'' plants, inhibiting the growth of untransformed plants very effectively. The neomycon phosphotransferase II gene is under control of the NOS promoter and terminator. |
==References:== | ==References:== | ||
| Line 90: | Line 94: | ||
<!-- Kopiervorlagen --> | <!-- Kopiervorlagen --> | ||
<!-- Ab hier richtige Referenzen einfügen --> | <!-- Ab hier richtige Referenzen einfügen --> | ||
| + | [[http://www.plant-biotech.net/paper/Reski_1998_BotActa-111_1_scan.pdf Reski, 1998]] Reski, R. (1998). Development, Genetics and Molecular Biology of Mosses. ''Bot. Acta'', 111:1-15.<br /> | ||
[[http://www.plantphysiol.org/content/127/4/1430 Schaefer and Zryd, 2001]] Schaefer, D.G. and Zrÿd, J. (2001). The Moss ''Physcomitrella patens'', Now and Then. ''Plant Physiology'', 127(4):1430-1438.<br /> | [[http://www.plantphysiol.org/content/127/4/1430 Schaefer and Zryd, 2001]] Schaefer, D.G. and Zrÿd, J. (2001). The Moss ''Physcomitrella patens'', Now and Then. ''Plant Physiology'', 127(4):1430-1438.<br /> | ||
| - | [[http://www. | + | [[http://www.ncbi.nlm.nih.gov/pubmed/14586556 Reski et al., 2004]] Hohe, A., Egener, T., Lucht, J.M., Holtorf, H., Reinhard, C., Schween, G. and Reski, R. (2004). An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. ''Curr Genet.'', 44(6):339-47.<br /> |
[[http://www.sciencedirect.com/science/article/pii/S0958166907000948 Reski and Decker, 2007]] Decker, E.L. and Reski, R. (2007). Moss bioreactors producing improved biopharmaceuticals. ''Current Opinion in Biotechnology'', 18(5):393-398.<br /> | [[http://www.sciencedirect.com/science/article/pii/S0958166907000948 Reski and Decker, 2007]] Decker, E.L. and Reski, R. (2007). Moss bioreactors producing improved biopharmaceuticals. ''Current Opinion in Biotechnology'', 18(5):393-398.<br /> | ||
| - | |||
| - | |||
[[http://www.ncbi.nlm.nih.gov/pubmed/22621344 Reski et al., 2012]] Parsons, J., Altmann, F., Arrenberg, C. K., Koprivova, A., Beike, A. K., Stemmer, C., Gorr, G., Reski, R. and Decker, E. L. (2012). Moss-based production of asialo-erythropoietin devoid of Lewis A and other plant-typical carbohydrate determinants. ''Plant Biotechnology Journal'', 10:851–861.<br /> | [[http://www.ncbi.nlm.nih.gov/pubmed/22621344 Reski et al., 2012]] Parsons, J., Altmann, F., Arrenberg, C. K., Koprivova, A., Beike, A. K., Stemmer, C., Gorr, G., Reski, R. and Decker, E. L. (2012). Moss-based production of asialo-erythropoietin devoid of Lewis A and other plant-typical carbohydrate determinants. ''Plant Biotechnology Journal'', 10:851–861.<br /> | ||
Latest revision as of 02:55, 29 October 2013
Physcomitrella - A new chassis for iGEM
General description
The moss Physcomitrella patens belongs to the land plant division Bryophyta, which are one of the earliest representatives of the land plants (Embryophyta) having evolved from green algae about 470 million years ago during the early Paleozoic. Hence mosses have a much simpler anatomy than higher land plants such as trees and flowering plants, which in particular means that they have not yet developed a vascular system, i.e an internal transport system for water and nutrients. Since they also lack a complex waterproofing system to prevent absorbed water from evaporating they need a moist environment to grow. Their main habitats are therefore shady and damp places such as woods and edges of streams but they are also found to be resistant to periods of drought and therefore can be found widely spread around the world, from the tropics to tundra regions, from coastal sand dunes up to high mountains.
The general organization of plant tissue into roots, stem and leaves is found in a much more basic version in mosses. They show a differentiated stem with simple leaves, usually only a single layer of cells thick and lacking veins, that are used to absorb water and nutrients. Instead of roots they have similar threadlike rhizoids http://www.plant-biotech.net/paper/Reski_1998_BotActa-111_1_scan.pdf Reski, 1998. These have a primary function as mechanical attachment rather than extraction of soil nutrients. Due to not having a vascular system bryophytes are doomed to stay small throughout their life-cycle typically stretching about 1-10 cm.
However different mosses and vascular plants are because of the early diverge of the evolutionary lineages, they share fundamental genetic and physiological processes. Hence a good approach to studying the complexity of higher land plants is to look at the bryophytes with their much simpler phenotype. Here researchers chose Physcomitrella patens as a model organism with a genome size of about 450 Mb along 27 chromosomes that is highly similar to other land plants in both exon-intron-structure and codon usage. http://www.plantphysiol.org/content/127/4/1430 Schaefer and Zryd, 2001
Life cycle
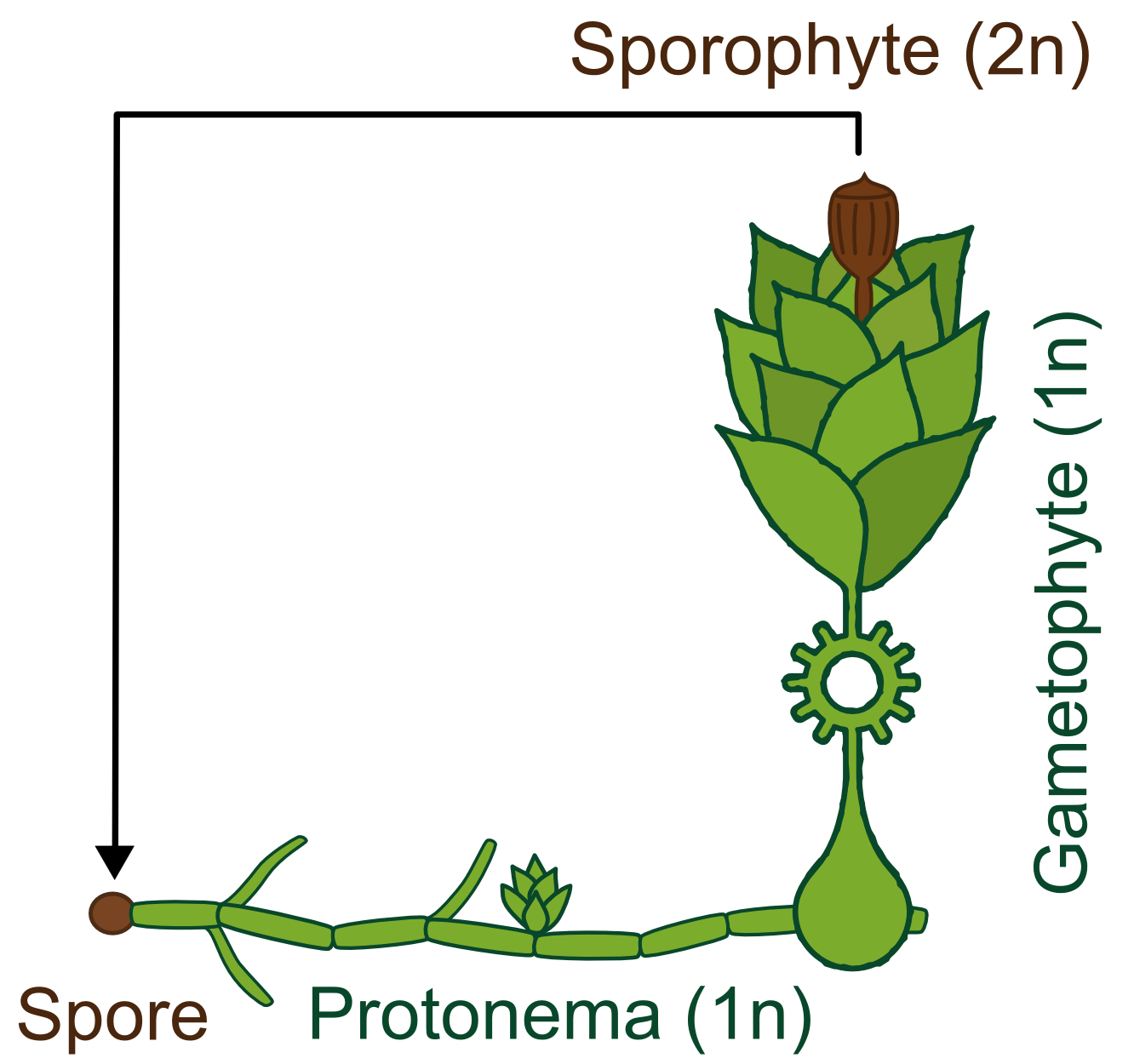
Generally land plants show an alternation of generations, the haploid (1n) gametophyte produces sperm and eggs which fuse and transform into the diploid (2n) sporophyte. This then forms haploid spores which become new gametophytes. Besides having no vascular system, bryophytes also differ from higher land plants in the fact that the gametophyte is the dominant phase of their life cycle, whereas in vascular plants the principal generation is the sporophyte.
The life cycle of P. patens http://www.plant-biotech.net/paper/Reski_1998_BotActa-111_1_scan.pdf Reski, 1998 only takes about 3 months and starts with the spore developing into a filamentous structure, the juvenile, transitory stage of the gametophyte, called protonema, which is composed of two types of cells. The chloronema cells with large and numerous chloroplasts mostly perform photosynthesis and thus supply the photoautotrophic plant with energy while the task of the caulonema cells is fast growth. The adult stage of the gametophyte, called gametophore ("gamete-bearer") has a more complex structure bearing leafs, stem and rhizoids. The transition from juvenile to adult gametophyte is started by initial cells in the protonema filament that differentiate into buds. The budding is therefore a single-cell-event, greatly stimulated by the plant hormone cytokinin, which promotes cell division.
The sex organs of the moss develop from the tip of the gametophore. P. patens is monoicous, meaning that male and female organs are produced in one plant. When liquid water is surrounding the tip, flagellate sperm cells can swim from the male sex organ to the female organ and fertilize the egg within. A zygote then develops into the sporophyte, which in turn produces thousands of haploid spores by meiosis. Sporophytes are typically physically attached to and dependent on supply from the dominating gametophyte.
Advantages of Physcomitrella as a model organism
General advantages
- P. patens stands out among the whole plant kingdom as the sole exception where gene targeting is feasible as an easy and fast routine procedure, even with an efficiency similar to S. cerevisiae, due to highly efficient homologous recombination. http://www.ncbi.nlm.nih.gov/pubmed/14586556 Reski et al., 2004 For that reason it is very easy to create knockout mosses by precise mutagenesis following the approach of reverse genetics in order to study the function of genes. Performing functional genomics in higher organisms is very important to understand biological functions of proteins in a multicellular context, e.g. in the context of cell-cell-contacts.
- As mosses mainly are in a haploid stage during their life cycle they are very straight-forward objects for genetics because complex backcrosses to determine changes in the genotype are not necessary
- Moss development starts with a filamentous tissue, the protonema, which is growing by apical cell division and, therefore is perfectly suitable for cell-lineage analysis since development of the plant can be pinpointed to the differentiation of a single cell. http://www.plant-biotech.net/paper/Reski_1998_BotActa-111_1_scan.pdf Reski, 1998 Also the simple life cycle makes P. patens a very useful item for developmental biology
- P. patens is increasingly used in biotechnology as a study object with implications for crop improvement or human health. Moss bioreactors (see Figure 2) can be used as an alternative to animal cell cultures (e.g. CHO cells) for the easy, inexpensive and safe production of complex biopharmaceuticals http://www.sciencedirect.com/science/article/pii/S0958166907000948 Reski and Decker, 2007 . For example it is a successful tool to produce asialo-EPO, a specific variant of Erythropoetin, which can perform its protective role by inhibiting apoptosis but has lost the potential doping activity. This safe drug is hard to produce in animal cell culture but easy to produce in the moss without impacting its growth or general performance http://www.ncbi.nlm.nih.gov/pubmed/22621344 Decker et al., 2012.
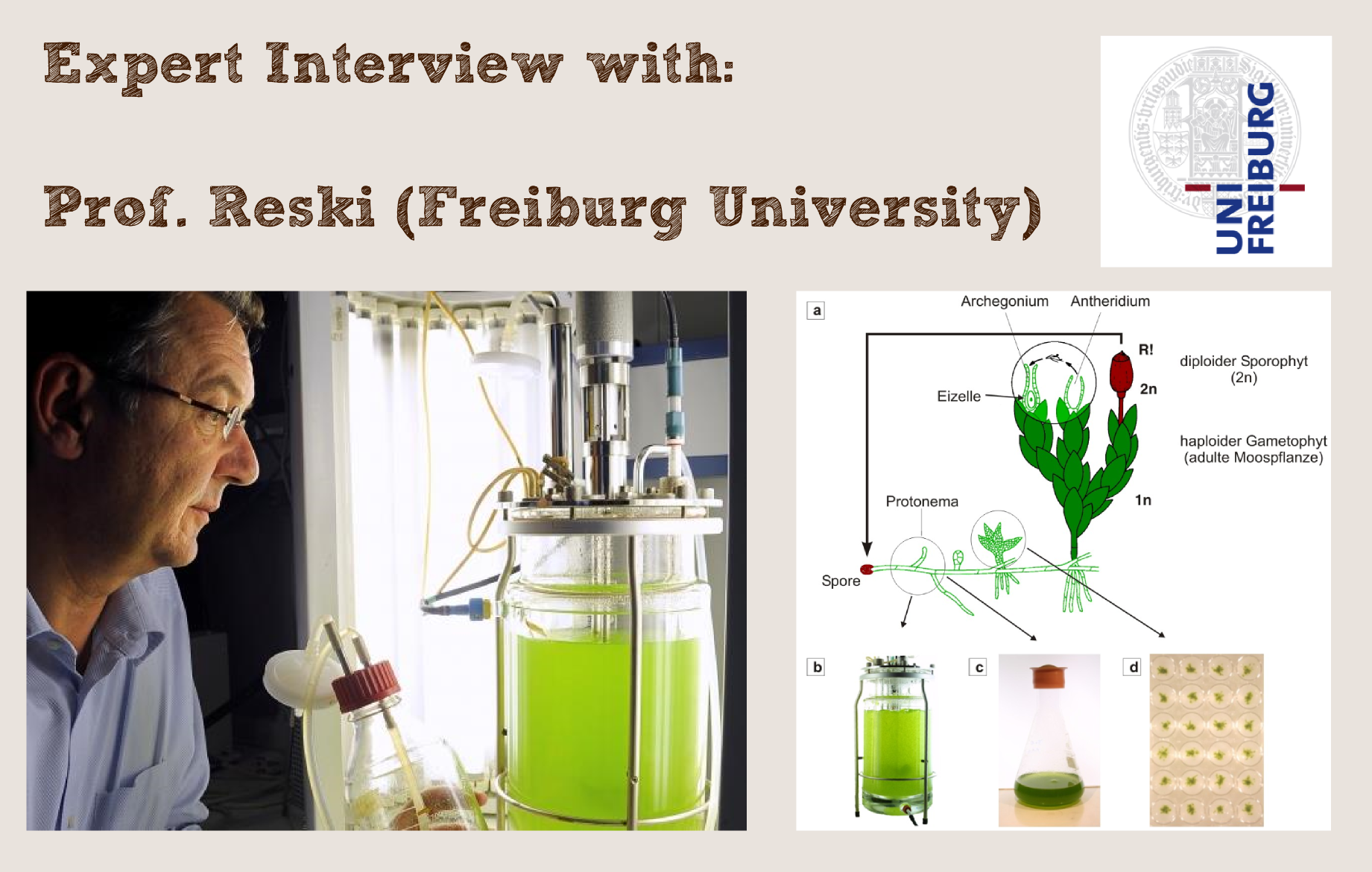
Expertbox: Prof. Reski
When we chose Phytoremediation as our project for this year´s competition, it soon became clear that Physcomitrella patens is a great chassis that could bring iGEM closer to the real world applications we were looking for. Therefore we contacted Prof. Dr. Reski, who is a recognized expert on Physcomitrella patens and its biotechnological applications.
Prof. Dr. Reski liked our idea of introducing his Physco to iGEM from the very beginning and offered his help wherever we would need it. Members of our team traveled to his lab in Freiburg (350 km) five times in total [1] to fetch plasmids known to be functional in Physcomitrealla patens or for discussions with him or his co-workers. We could win him over as an advisor for our team, which shows how iGEM brings together universities and scientific groups from many different places.
Advantages as a new chassis for iGEM
- As a plant, P. patens offers interesting opportunities for application as it is self sustaining, renewable and a natural part of our environment. Therefore it is much easier to implement it into real world scenarios than bacteria or yeast. And although there is the disadvantage of having to wait about 4-6 weeks after transformation until experiments can be done, this is still very short considering the high complexity of the organism. Working with P. patens can easily be done in the timeframe of the competition by preparing the DNA constructs in bacteria.
- P. patens is a well studied model organism which means that besides having its full genome sequenced in 2006 there are well equipped [http://www.cosmoss.org databases]. Furthermore, there exists an [http://www.moss-stock-center.org International Moss Stock Center (IMSC)] in which many ecotypes, mutants and transgenic strains of P. patens are stored and accessible to the scientific community. So there is enough knowledge and material to work on for synthetic biologists.
- At the same time the moss offers access to very exciting new physiological processes since it is a much more complex multicellular eukaryotic organism than the chassis already established in iGEM.
- P. patens is an easy plant to work with and requires neither expensive maintenance facilities nor large laboratory space. Most of the basic tools for high precision mutagenesis have been tested on this plant, were found to work and are easily available (see Moss methods below).
Moss methods - working with Physcomitrella
The techniques used for the cultivation and manipulation of Physcomitrella patens are based on the knowledge of the chair for Plantbiotechnology of Prof. Dr. Reski at Freiburg University, which are available at [http://www.plant-biotech.net Plant-Biotech.net].
- Cell culture: Physcomitrella patens plants can be cultivated either on solidified medium or in liquid medium. Liquid cultures can be kept in Erlenmeyer flasks under constant rotation and light exposure or in bioreactors for large scale production. By regularly (ideally weekly) disrupting the plants mechanically with a mixer (e.g. Ultra-Turrax) a homogenized culture can be achieved.
- Storage: Long-term storage of Physcomitrella strains can be achieved by cryo-preservation. This procedure ensures a maximum survival rate of the plant by preconditioning and controlled freezing. http://www.plant-biotech.net/paper/PlantBiol_2004_Schulte_pageproof.pdf Schulte and Reski, 2004
- Targeted knockout: P. patens is unique among plants in its high efficiency of gene targeting by homologous recombination. To knockout a specific gene a disruption construct has to be generated. This consists of the gene to be silenced with a selection cassette (usually the nptII gene) inserted into its center by suitable restriction sites. The moss is then transformed with this DNA construct which is integrated into the genome by homologous recombination. http://www.ncbi.nlm.nih.gov/pubmed/14586556 Reski et al., 2004
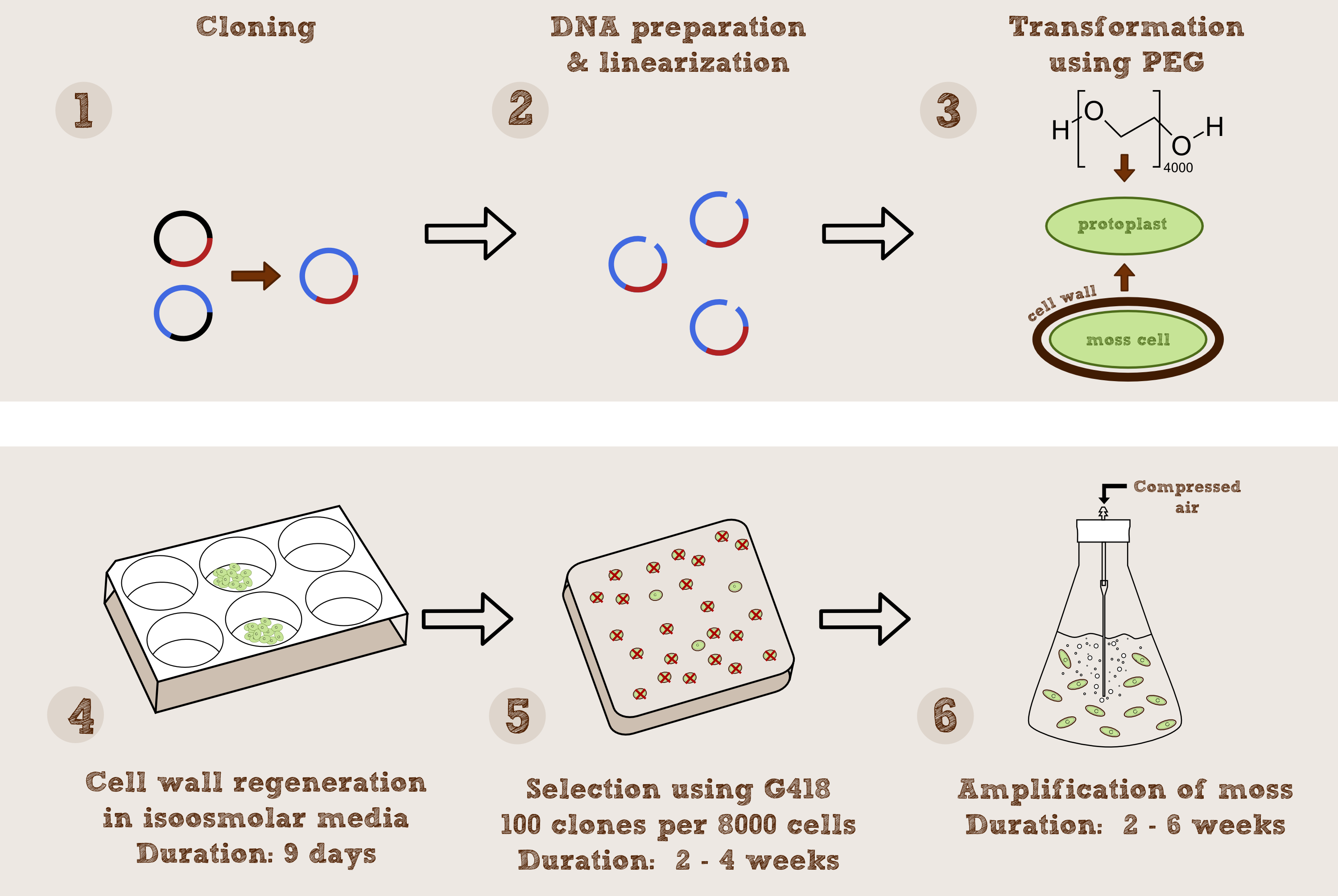
- Transformation: Transformation of P. patens requires protoplasts of the plant, which can be obtained by cell wall digesting enzymes. The protoplasts can either be transformed by particle bombardment or via polyethylene glycol (PEG), which is the easiest and most commonly used method. The DNA constructs should be linearized for optimal transformation and contain a selection marker for subsequent screening. After transformation, the protoplasts are cultivated in the dark for 12-16 h followed by 9 days under normal growth conditions during which they regenerate their cell wall. Afterwards they are plated on solidified medium in normal petri dishes and first experiments can be executed. http://link.springer.com/article/10.1007/BF00260654 Schaefer et al., 1991
- Analysis of transformants: First, the transformed protoplasts are plated on solidified medium containing the antibiotic G418 (Geneticin), to which successfully transformed plants are resistant due to the selection marker nptII which encodes the enzyme neomycin phosphotransferase. Secondly, the transformants are analyzed by a PCR screen using primers derived the selection marker cassette´s sequences http://www.plant-biotech.net/paper/PlantMolecularBiologyReporter_2002_Schween.pdf Schween et al., 2002.
In conclusion, it naturally takes a bit more time to work with a plant in comparison to bacteria or yeast (see figure 6), but the advantages definitely outweigh the time factor and if teams start early enough, it is perfectly possible to work with P. patens as a chassis in an iGEM project!
This is a brief summary of the most important techniques in working with P. patens. For a full description of these methods please see our methods page.
BioBricks for protein expression in Physcomitrella
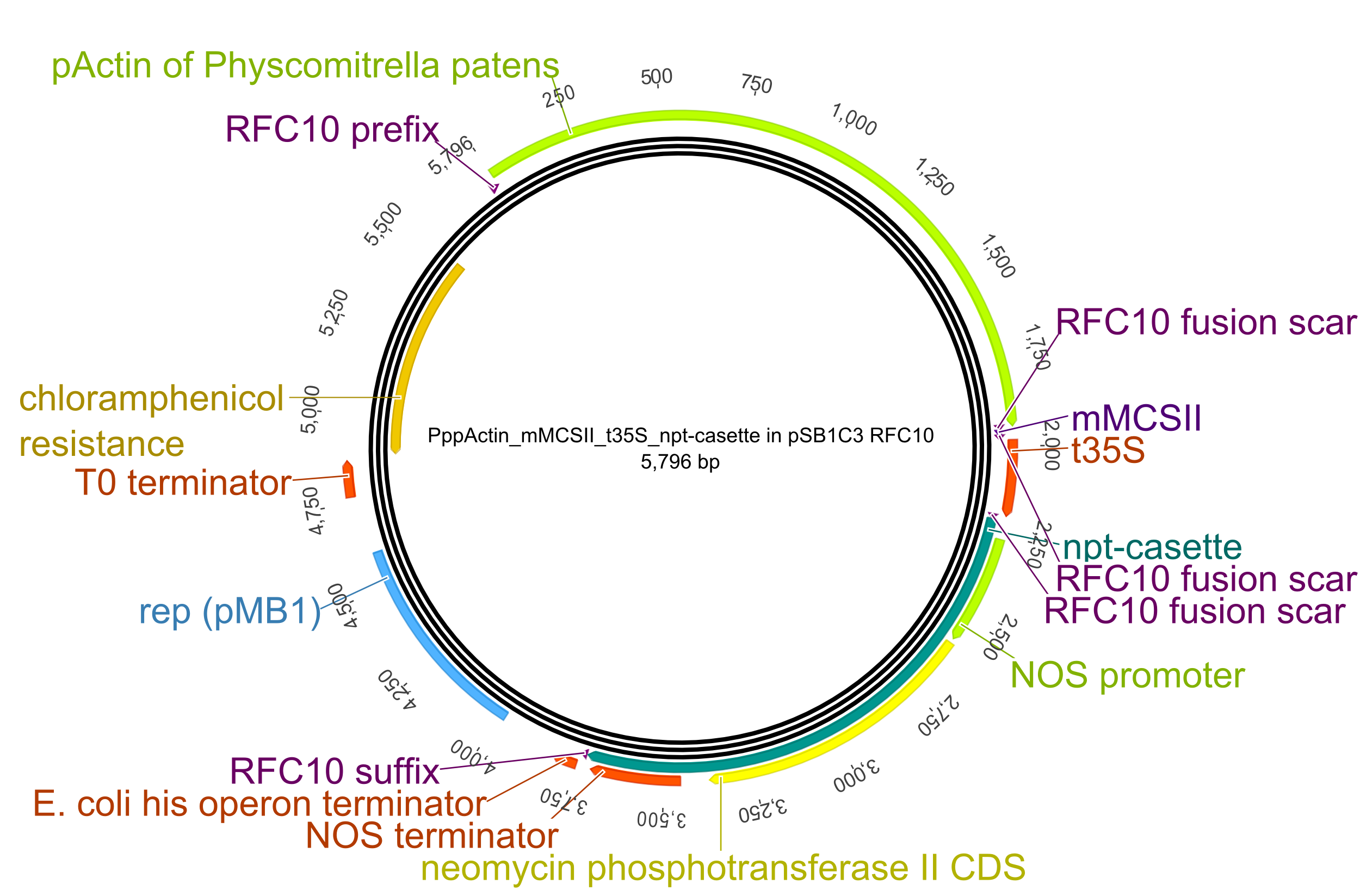
To transform the moss, we created an all-purpose backbone into which all our constructs could be integrated easily. Therefore we modified the common iGEM pSB1C3 plasmid with the following plant-specific components:
- pAct5 (Actin 5 promoter from Physcomitrella patens): for strong protein expression.
- miniMCS: This miniature multiple cloning site contains restriction sites for the enzymes MfeI and SbfI which allows you to insert BioBrick between two other Biobricks (in our case: pAct5 and t35S_npt-cassette) as a result of the compability of MfeI with EcoRI and SbfI with PstI.
- t35S (35S terminator): This terminator originates from the 35S transcript of the cauliflower mosaic virus.
- nptII-cassette: This selection cassette contains the neomycon phosphotransferase II gene, encoding the enzyme aminoglycoside-3'-phosphotransferase (NPTII), which inactivates a range of aminoglycoside antibiotics such as Kanamycin, Neomycin and Geneticin (G418) by phosphorylation. The latter antibiotic is used as selective agent for transformed P. patens plants, inhibiting the growth of untransformed plants very effectively. The neomycon phosphotransferase II gene is under control of the NOS promoter and terminator.
References:
http://www.plant-biotech.net/paper/Reski_1998_BotActa-111_1_scan.pdf Reski, 1998 Reski, R. (1998). Development, Genetics and Molecular Biology of Mosses. Bot. Acta, 111:1-15.
http://www.plantphysiol.org/content/127/4/1430 Schaefer and Zryd, 2001 Schaefer, D.G. and Zrÿd, J. (2001). The Moss Physcomitrella patens, Now and Then. Plant Physiology, 127(4):1430-1438.
http://www.ncbi.nlm.nih.gov/pubmed/14586556 Reski et al., 2004 Hohe, A., Egener, T., Lucht, J.M., Holtorf, H., Reinhard, C., Schween, G. and Reski, R. (2004). An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. Curr Genet., 44(6):339-47.
http://www.sciencedirect.com/science/article/pii/S0958166907000948 Reski and Decker, 2007 Decker, E.L. and Reski, R. (2007). Moss bioreactors producing improved biopharmaceuticals. Current Opinion in Biotechnology, 18(5):393-398.
http://www.ncbi.nlm.nih.gov/pubmed/22621344 Reski et al., 2012 Parsons, J., Altmann, F., Arrenberg, C. K., Koprivova, A., Beike, A. K., Stemmer, C., Gorr, G., Reski, R. and Decker, E. L. (2012). Moss-based production of asialo-erythropoietin devoid of Lewis A and other plant-typical carbohydrate determinants. Plant Biotechnology Journal, 10:851–861.
http://www.plant-biotech.net/paper/PlantBiol_2004_Schulte_pageproof.pdf Schulte and Reski, 2004 Schulte, J. and Reski, R. (2004). High throughput Cryopreservation of 140 000 Physcomitrella patens Mutants. Plant Biology, 6:119-127.
http://link.springer.com/article/10.1007/BF00260654 Schaefer et al., 1991 Schaefer, D., Zryd, J.-P., Knight, C., Cove, D. (1991). Stable transformation of the moss Physcomitrella patens. Molecular and General Genetics, 226:418-424.
http://www.plant-biotech.net/paper/PlantMolecularBiologyReporter_2002_Schween.pdf Schween et al., 2002 Schween, G., Fleig, S., Reski, R. (2002). High-throughput-PCR screen of 15,000 transgenic Physcomitrella plants. Plant Molecular Biology Reporter, 20:43–47.
 "
"










AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351