Team:British Columbia/Project/CRISPR
From 2013.igem.org
AnnaMüller (Talk | contribs) |
AnnaMüller (Talk | contribs) |
||
| (60 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:British_Columbia/Templates/MainHeader}} | {{:Team:British_Columbia/Templates/MainHeader}} | ||
<html> | <html> | ||
| + | <a name="top"></a> | ||
<style> | <style> | ||
.icon { width: 210px; margin 5px;} | .icon { width: 210px; margin 5px;} | ||
| Line 35: | Line 36: | ||
<div id="frontpage"> | <div id="frontpage"> | ||
<img id="frontpic" src="https://static.igem.org/mediawiki/2013/1/1b/UBC-CRISPR-Mechanism-Out.png"> | <img id="frontpic" src="https://static.igem.org/mediawiki/2013/1/1b/UBC-CRISPR-Mechanism-Out.png"> | ||
| + | |||
</div> | </div> | ||
</html> | </html> | ||
| + | =CRISPR= | ||
===What are CRISPRs and how do they work?=== | ===What are CRISPRs and how do they work?=== | ||
| - | CRISPRs ('''C'''lustered '''R'''egularly '''I'''nterspaced '''S'''hort '''P'''alindromic '''R'''epeats) are | + | CRISPRs ('''C'''lustered '''R'''egularly '''I'''nterspaced '''S'''hort '''P'''alindromic '''R'''epeats) are specific regions in some bacterial and archaeal genomes that, together with associated Cas ('''C'''RISPR-'''as'''sociated) genes, function as an adaptive immune system in prokaryotes. While the specific ‘adaptive’ nature of this immunity is still under investigation, it is known that exogenous DNA is processed by Cas proteins into short (~30 base pair) sequences that are adjacent to the '''P'''rotospacer '''A'''djacent '''M'''otif (PAM) site. These short pieces of DNA are then incorporated into the host genome between repeat sequences to form '''spacer''' elements. The repeat-spacer-repeat array is constitutively expressed (pre-CRISPR RNAs or pre-crRNAs) and processed by Cas proteins to form small RNAs (crRNAs). The small RNAs are then loaded into Cas proteins and act to guide them to initiate the sequence-specific cleavage of the target sequence. |
===What did we do with them?=== | ===What did we do with them?=== | ||
| - | As an adaptive immune response, we wanted to know if the CRISPR system could be put together in a modular way to confer resistance to known phage genomes | + | As an adaptive immune response, we wanted to know if the CRISPR system could be put together in a modular way to confer resistance to known phage genomes, thereby vaccinating the host. We first wrote scripts capable of '''identifying the most [[#Spacer Selection|broadly neutralizing spacer regions]]''' from compiled phage genomes. We then '''assembled the minimum CRISPR components into [[#System Design|BioBricks]]''' and conducted the necessary '''[[#Results|proof-of-concept experiments]] in <i>''E. coli''.'''</i> First, we characterized the dynamics of phage infection in our specific host strain and experimental protocols. We then built a system that protects E.coli against T4 phage infection and are beginning to understand some guidelines for assembling CRISPR components to provide immunity. Currently, we are carrying out further validation experiments, extending our system to work with T7 phage, performing some <i>in vitro</i> characterizations, and exploring new possibilities with our working and the tunable CRISPR system. |
| - | === | + | ===Why do this?=== |
| - | Ultimately, we hope that large-scale fermenters could be vaccinated | + | Ultimately, we hope that large-scale fermenters could be vaccinated to prevent bacterial population collapse caused by environmental phage infection. To extend the application of this approach, we designed specific neutralization elements that allow for population level programming of a culture. We envisioned this first being applied to yogurt where the biosynthesis of flavour combinations might be controlled by targeted phage addition. Check out our [[Team:British_Columbia/Project/PopulationControl|population control]] section for further details on the construction of a few of these pathways. Approaching this from another angle, our [[Team:British_Columbia/Modeling|modeling]] section aims to describe the population dynamics of both vulnerable and immunized populations under phage predation with the hopes of eventually being able to rationally tune relative proportions of flavour compounds. |
==System Design== | ==System Design== | ||
| - | The three stages of CRISPR immunity | + | The three stages of CRISPR immunity (spacer acquisition, crRNA biogenesis, and target interferences), and the general gene architecture described above, are hallmarks of the CRISPR/Cas system. There is, however, substantial diversity in the naturally occurring spacer elements, the Cas genes and the functional PAM sequences. Approximately 45 different gene families have been identified, which, when combined with analysis of their repeat and PAM sequences, can lead to their categorization into three main groups (Types I, II and III) with 10 subtypes (IA, IB, IC, ID, IE, IF, IIA, IIB, IIIA and IIIB). |
| - | We chose to work with the type IIA CRISPR3 locus from Streptococcus thermophilus because it has been previously shown to confer immunity to double stranded DNA (dsDNA) phage and plasmids in ''Escherichia coli'' and it requires a minimum set of only three | + | We chose to work with the type IIA CRISPR3 locus from <i>Streptococcus thermophilus</i> because it has been previously shown to confer immunity to double stranded DNA (dsDNA) phage and plasmids in <i>''Escherichia coli''</i> and it requires a minimum set of only three components—one protein (Cas9), a non-coding small RNA (trans-activating crRNA, tracrRNA), and the repeat-spacer array. These three necessary components were decoupled from their native regulatory framework and assembled. |
<html> | <html> | ||
| Line 63: | Line 66: | ||
===Cas9=== | ===Cas9=== | ||
| - | Cas9 is the hallmark of the type II | + | Cas9 is the hallmark of the type II system—a massive 163 kDa multifunctional protein involved in the processing of pre-crRNA and the cleavage of exogenous DNA. The <i>Cas9</i> gene was subcloned out of [http://www.addgene.org/39314/ pMJ824] and assembled under both the arabinose inducible pBAD [http://parts.igem.org/Part:BBa_I13453 BBa_I13453] and constitutive [http://parts.igem.org/Part:BBa_J23118 BBa_J23118] promoters. Cas9 was submitted to the registry as part [http://parts.igem.org/Part:BBa_K1129006 BBa_K1129006]. Expression of Cas9 under both arabinose induction and constitutive expression was confirmed with SDS-PAGE. |
===Trans-activating crRNA=== | ===Trans-activating crRNA=== | ||
| - | The tracrRNA is a 36 base, small non-coding RNA of which 25 bases is complementary to the repeat sequence. The hybridization of the tracrRNA to the repeat sequence in the pre-crRNA results in double-stranded RNA that is recognized and cleaved by a non-Cas RNaseIII enzyme. It has also been shown that tracrRNA, the mature crRNA and Cas9 are all required for cleavage of target dsDNA. | + | The tracrRNA is a 36-base, small, non-coding RNA sequence of which 25 bases is complementary to the repeat sequence. The hybridization of the tracrRNA to the repeat sequence in the pre-crRNA results in double-stranded RNA that is recognized, and cleaved by, a non-Cas RNaseIII enzyme. It has also been shown that tracrRNA, the mature crRNA and Cas9 are all required for cleavage of target dsDNA. |
The tracrRNA was cloned from annealed oligonucleotides and assembled under the arabinose inducible pBAD [http://parts.igem.org/Part:BBa_I13453 BBa_I13453] promoter. It was submitted to the registry as parts [http://parts.igem.org/Part:BBa_K1129008 BBa_K1129008] (sense) and [http://parts.igem.org/Part:BBa_K1129009 BBa_K1129009] (anti-sense). | The tracrRNA was cloned from annealed oligonucleotides and assembled under the arabinose inducible pBAD [http://parts.igem.org/Part:BBa_I13453 BBa_I13453] promoter. It was submitted to the registry as parts [http://parts.igem.org/Part:BBa_K1129008 BBa_K1129008] (sense) and [http://parts.igem.org/Part:BBa_K1129009 BBa_K1129009] (anti-sense). | ||
===Spacer Selection=== | ===Spacer Selection=== | ||
| - | Spacers were selected using a perl script (example [https://static.igem.org/mediawiki/2013/1/13/UBC_spacer_selection.zip here]) which searches a set of viral genomes to find any occurrences of the PAM site <tt>NGGNG</tt>. Spacer sequences were removed from the set if 1) the 9 bases preceding the PAM were also found adjacent to a PAM site in the host genome, or 2) if they contained illegal restriction sites. For the remaining spacers, the 30 bases prior to the PAM site were recorded and their frequency of occurrence in each genome was determined. | + | Spacers were selected using a perl script (example [https://static.igem.org/mediawiki/2013/1/13/UBC_spacer_selection.zip here]) which searches a set of viral genomes to find any occurrences of the PAM site <tt>NGGNG</tt>. Spacer sequences were removed from the set if (1) the 9 bases preceding the PAM were also found adjacent to a PAM site in the host genome, or (2) if they contained illegal restriction sites. For the remaining spacers, the 30 bases prior to the PAM site were recorded and their frequency of occurrence in each genome was determined. |
<center> | <center> | ||
| Line 79: | Line 82: | ||
</center> | </center> | ||
| - | We reasoned | + | We reasoned the spacers that are the most heavily conserved among related genomes would be under the greatest selective pressure to be retained and would provide a wider range of immunity. Based on this, we selected the spacers which were the most widely distributed in the T7, T4, and closely related genomes for our system. By calculating the fraction of the phage genomes that would be neutralized by combining the most widely distributed spacers, we showed that using our approach only 5 and 3 spacers would be required to neutralize all of the phage related to T4 and T7, respectively. |
===Toxic Plasmids=== | ===Toxic Plasmids=== | ||
| Line 89: | Line 92: | ||
</html> | </html> | ||
| - | In order to assay for CRISPR-mediated defence against plasmids, we designed two plasmids | + | In order to assay for CRISPR-mediated defence against plasmids, we designed two plasmids that, when cleaved, will eliminate the antibiotic resistance conferred by the “invading” plasmid, thereby resulting in cell death. These two plasmids contain the spacer and PAM sequence identified by our informatics pipeline and an ampicillin resistance marker. They have been submitted to the registry (with chloramphenicol resistance) in parts [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1129010 BBa_K1129010] (T4) and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1129011 BBa_K1129011] (T7). |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<p> | <p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 98: | Line 110: | ||
====Normal growth under T4 Phage Infection==== | ====Normal growth under T4 Phage Infection==== | ||
| - | Our first goal was to measure bacterial growth kinetics under T4 phage predation with our host strain and experimental conditions. The | + | Our first goal was to measure bacterial growth kinetics under T4 phage predation with our host strain and experimental conditions. The upper left quadrant has the estimated inoculums and four different dilutions of phage were assessed. The results are consistent with a step-wise lysis cycle that is well documented in the literature. See the [[Team:British_Columbia/Modeling|modeling section]] for more detailed analysis of the kinetics. |
<center> | <center> | ||
[[File:UBC-Growth_curves_MOI.png|800px]] | [[File:UBC-Growth_curves_MOI.png|800px]] | ||
</center> | </center> | ||
| - | '''Figure 4.''' The effect of initial phage concentration on E.coli GB10 cell growth for various inoculums. Overnight cultures of wild-type ''E. coli'' 10G cells were grown at 37 °C until an OD of 0.3 was obtained diluted to each inoculum, the cells counts of which were estimate by plate counts (CFU). The cultures were grown in a 96-well format and OD was measured every 5 min for 2 hrs (n = 2). | + | '''Figure 4.''' The effect of initial phage concentration on <i>''E. coli''</i> GB10 cell growth for various inoculums. Overnight cultures of wild-type <i>''E. coli''</i> 10G cells were grown at 37 °C until an OD of 0.3 was obtained diluted to each inoculum, the cells counts of which were estimate by plate counts (CFU). The cultures were grown in a 96-well format and OD was measured every 5 min for 2 hrs (n = 2). |
====Combinatorial CRISPR-Cas9 element assembly==== | ====Combinatorial CRISPR-Cas9 element assembly==== | ||
| - | Once we were confident in our ability to measure growth under phage predation, we designed a strategy to assemble Cas9, tracrRNA (sense/antisense) and the T4 spacer-repeat-spacer with inducible promoters in a combinatorial pattern and made a library of approximately 800 clones (see protocols). We then added 10<sup>3</sup> phage to each clone and screened for growth in high-throughput. | + | Once we were confident in our ability to measure growth under phage predation, we designed a strategy to assemble Cas9, tracrRNA (sense/antisense), and the T4 spacer-repeat-spacer with inducible promoters in a combinatorial pattern and made a library of approximately 800 clones (see protocols). We then added 10<sup>3</sup> phage to each clone and screened for growth in high-throughput. |
<center> | <center> | ||
[[File:UBC-CRISPR-Screen-Scatter.png|650px]] | [[File:UBC-CRISPR-Screen-Scatter.png|650px]] | ||
| Line 119: | Line 131: | ||
====Growth kinetics of immunized culture==== | ====Growth kinetics of immunized culture==== | ||
| - | From the combinatorial library, we performed colony PCR on 30 clones and showed that the majority of constructs had an approximately 5 kb long insert. A subset of these, along with the clones selected in Figure 2 were arrayed into a 96-well plate. We then added 10<sup>3</sup> T4 phage to each well and measured OD in the same way as in Figure 1. This seemed to show that there were two distinct groups, one with increased fitness in the presence of phage. | + | From the combinatorial library, we performed colony PCR on 30 clones and showed that the majority of constructs had an approximately 5 kb long insert. A subset of these, along with the clones selected in Figure 2, were arrayed into a 96-well plate. We then added 10<sup>3</sup> T4 phage to each well and measured OD in the same way as in Figure 1. This seemed to show that there were two distinct groups, one with increased fitness in the presence of phage. |
<center> | <center> | ||
[[File:UBC_Library_Growth.png|600px]] | [[File:UBC_Library_Growth.png|600px]] | ||
</center> | </center> | ||
| - | '''Figure 7.''' The effect of phage predation on clones from combinatorial libraries. Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 10<sup>3</sup> phage were then added to each well and OD was monitored overnight. The results seem to resolve two groups, one showing increased final OD over the other (blue and red lines respectively (n=2). | + | '''Figure 7.''' The effect of phage predation on clones from combinatorial libraries. Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 10<sup>3</sup> phage were then added to each well and OD was monitored overnight. The results seem to resolve two groups, one showing increased final OD over the other (blue and red lines respectively) (n=2). |
| Line 130: | Line 142: | ||
[[File:UBC-CRISPR-Growth.png|650px]] | [[File:UBC-CRISPR-Growth.png|650px]] | ||
</center> | </center> | ||
| - | '''Figure 8.''' Monitoring growth of two select combinatorial library clones under high cell inoculum. 10 | + | '''Figure 8.''' Monitoring growth of two select combinatorial library clones under high cell inoculum. 10<sup>3</sup> phage were then added to each well and OD was monitored overnight (n=2). |
| + | |||
| + | ====Testing host background dependance==== | ||
| + | |||
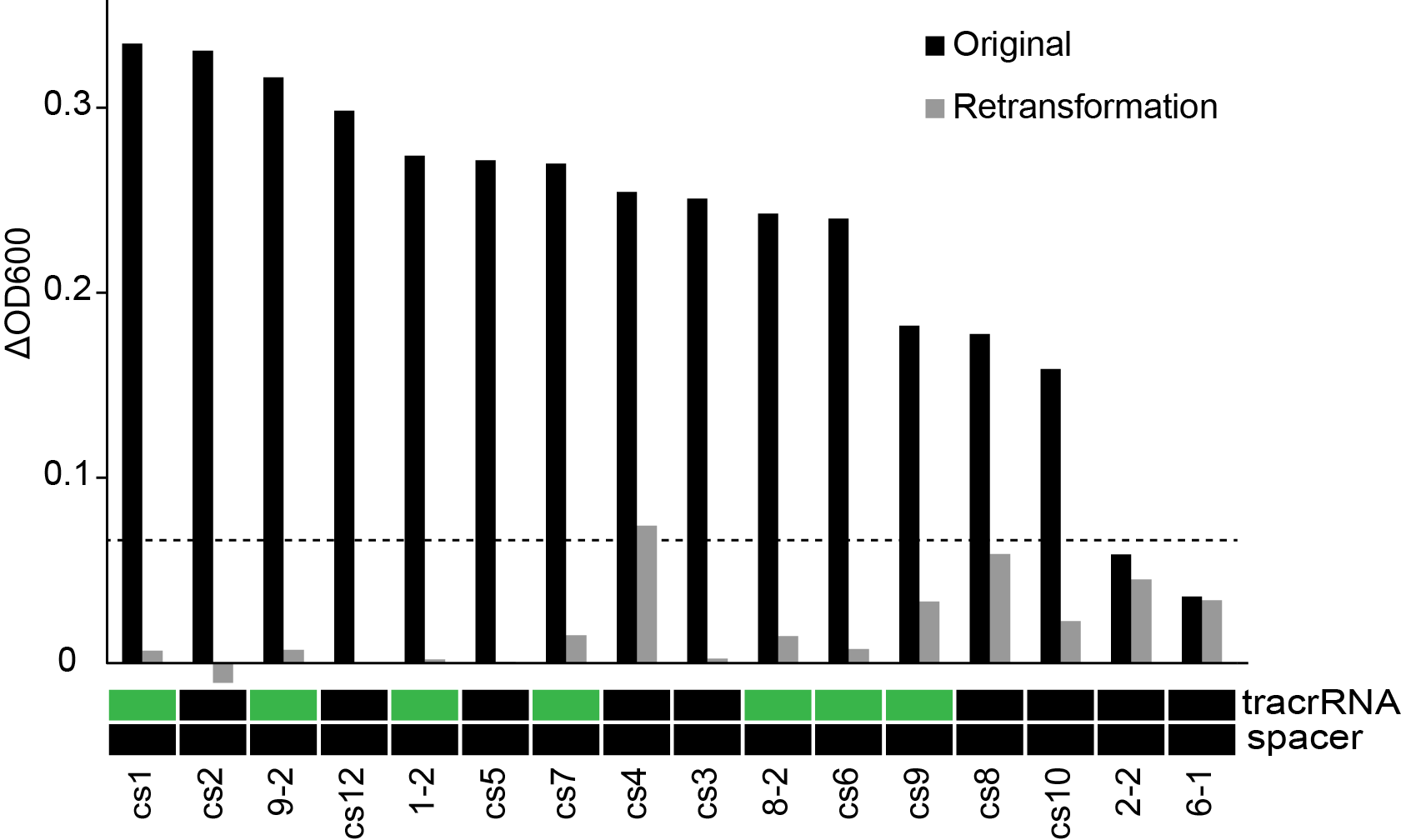
| + | To ensure that the survival phenotype we were observing was in-fact dependant on the expected constructs, we re-isolated plasmid from "survivors" and re-assayed their growth under phage predation. We also sent these plasmids for sequencing as to assign a genotype that resulted from our combinatorial assembly. In a new host background, the growth advantage was not maintained, suggesting that the phenotype was dependant on changes occurring in the host background (Figure 9). Furthermore, the plasmid sequences showed that the repeat-spacer regions was not present, showing that the combinatorial screening strategy did now work as expected. There could, however, still be something interesting happening where the host strain is able to acquire resistance more readily while expressing Cas9. There was also only a slight correlation between the presence of tracrRNA and survival. We are looking at this more closely. More importantly, we need to be careful looking using high-throughput approaches as this increases the rate at which spontaneous processes can confound the results. | ||
| + | <center> | ||
| + | [[File:UBC_CRISPR_ScreenGT.png|700px]] | ||
| + | </center> | ||
| + | '''Figure 9.''' The effect of phage predation on clones from combinatorial libraries after re-transformation and genotyping. Change in OD in culture in a 96-well plate grown for 11 hours at 30 °C. 10<sup>3</sup> phage were then added to each well and OD was monitored overnight (n=3). Clones containing either tracrRNA or spacers are shown in green. | ||
| + | |||
| + | ====Testing the standard assembled construct==== | ||
| + | |||
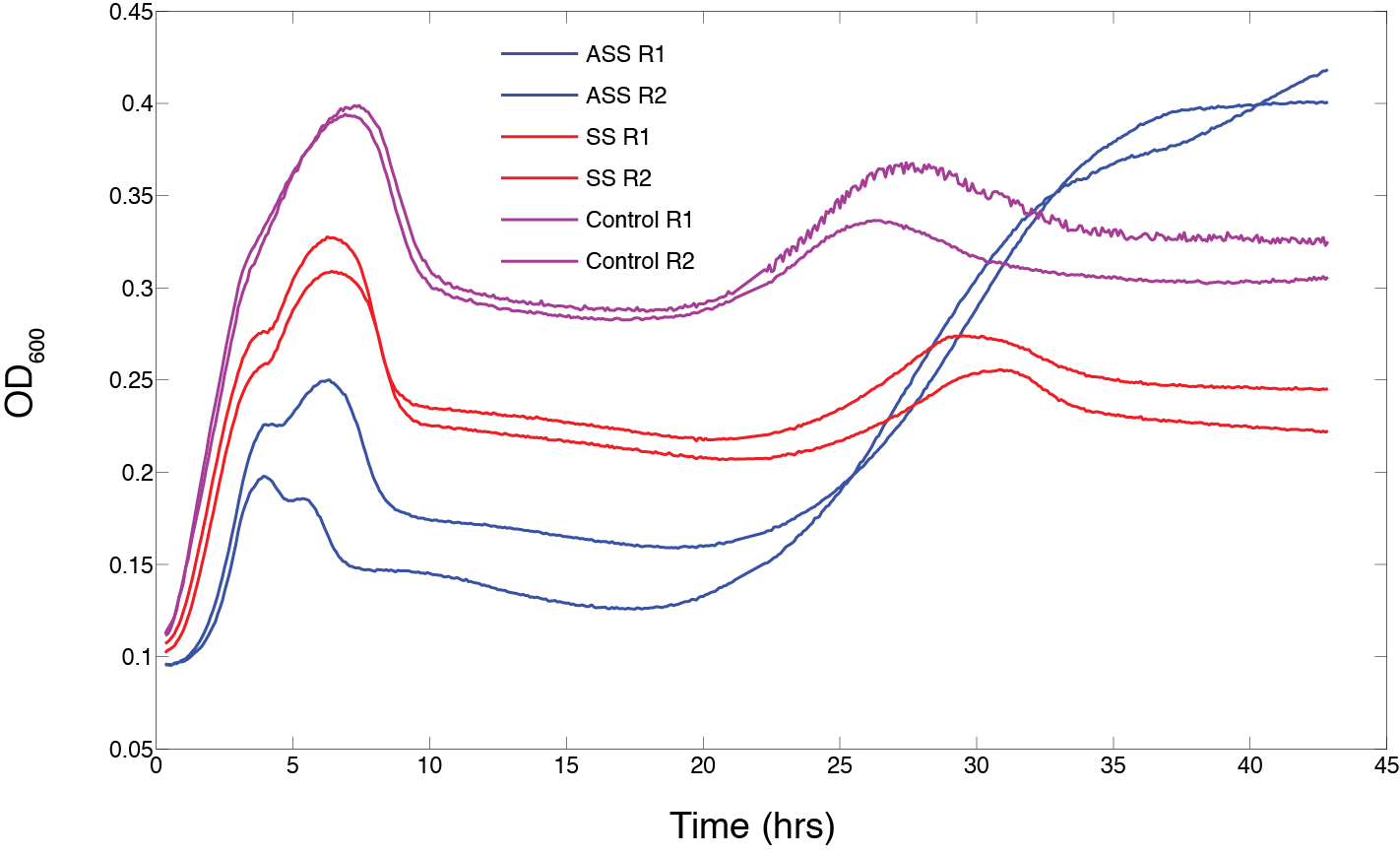
| + | We then finished the standard assembly of our designed CRISPR construct and tested growth under T4 phage predation. The construct contained cas9, the repeat-spacer region and both orientations of the tracrRNA. This showed an interesting phenotype where the entire construct seems to cause enhanced recovery after a cycle of phage predation (starting at approximately 25 hrs). The initial infection cycle (from 0 to 15 hrs) showed that cas9 caused a significant hindrance in growth compared to an empty vector and the repeat-spacer/tracrRNA control (Figure 9). | ||
| + | <center> | ||
| + | [[File: Growth_retrans.png|650px]] | ||
| + | </center> | ||
| + | '''Figure 9.''' The effect of phage predation on clones from assembled CRISPR constructs all under inducible promoters: cas9/repeat-spacer/tracrRNA-sense (AAS), repeat-spacer/tracrRNA-sense (SS), and empty vector (control). Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 10<sup>3</sup> phage and 1 ug/ml of arabinose was then added to each well and OD was monitored for 46 hrs (n=2). | ||
| + | |||
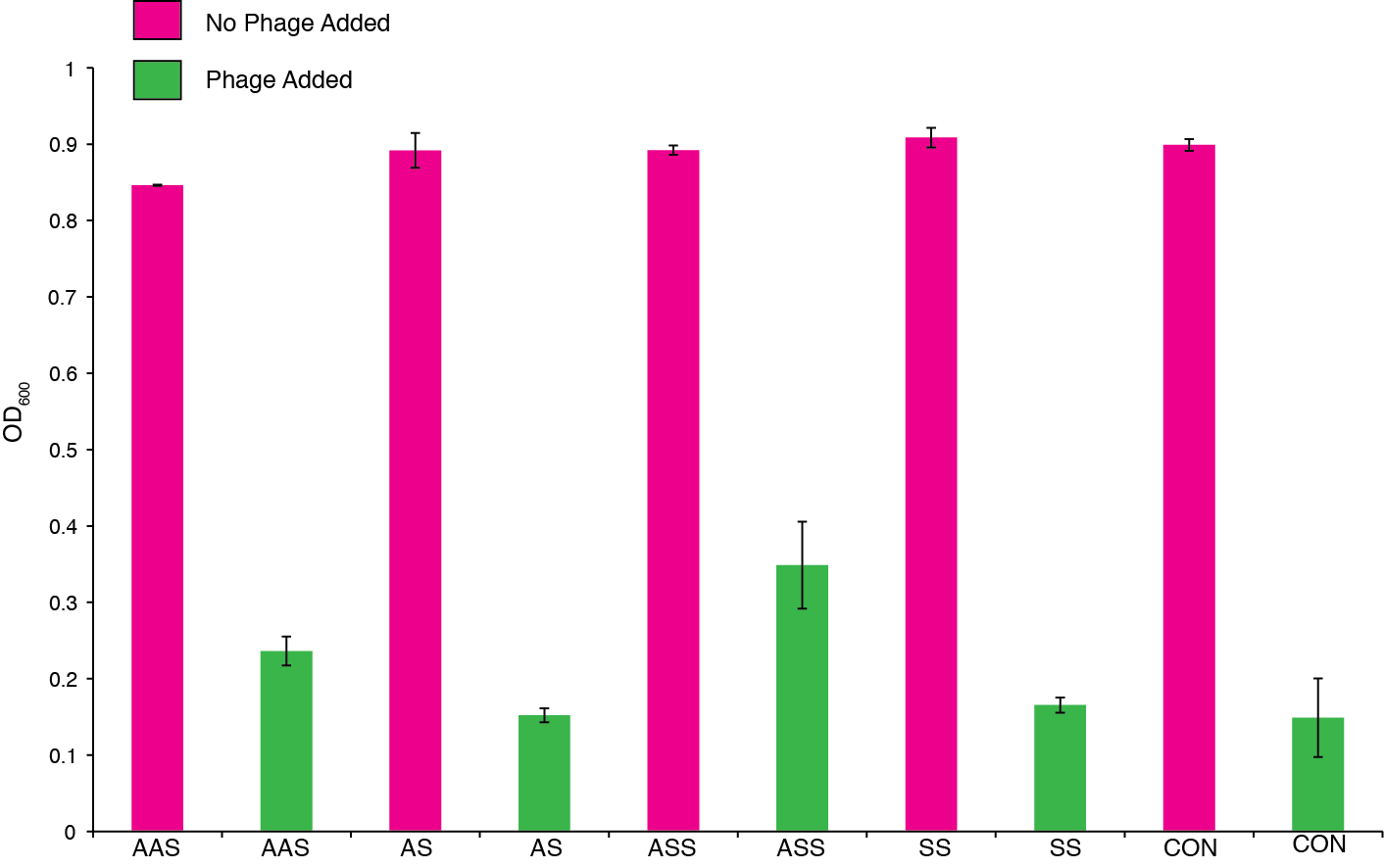
| + | We then monitored this trend by taking final OD after 46 hrs and ensured that the phenotype was consistent under various arabinose and phage concentrations. Figure 10 shows final OD with a higher phage inoculum. | ||
| + | <center> | ||
| + | [[File: Final_OD.png|650px]] | ||
| + | </center> | ||
| + | '''Figure 10.''' The effect of phage predation on clones from assembled CRISPR constructs all under inducible promoters: cas9/repeat-spacer/tracrRNA-sense (ASS), repeat-spacer/tracrRNA-sense (SS), and empty vector (control). Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 10<sup>4</sup> phage and 1 ug/ml of arabinose was then OD was measured after 46 hrs. | ||
| + | <br> | ||
| + | |||
| + | ====Transformation assay==== | ||
| + | |||
| + | While the phage phenotypes show promise for providing a growth advantage under phage, we also wanted to show some evidence for the sequence specificity of our CRISPR construct. The experimental approach is also described above in the toxic plasmids section. We designed a transformation assay which mixed a RFP containing control plasmid and a decoy plasmid which contained the targeted T4 and PAM sequence (both on PSB1A3, to be maintained along with PSB1C3 under both ampicillin and chloramphenicol selection). We would then expect that if the decoy plasmid was being cleaved, the ratio of red/white colonies counted on a plated would increase as the decoy plasmid was being cleaved. This was in-fact observed for the entire construct, and controlled for using a construct that was missing Cas9 (Figure 11). | ||
| + | <center> | ||
| + | [[File: Oct22_Transformation_Asssay.png|650px]] | ||
| + | </center> | ||
| + | '''Figure 11.''' Colony counts of RFP and decoy plasmid containing clones (red and white, respectively): Cas9/repeat-spacer/tracrRNA-sense (CSS) and repeat-spacer/tracrRNA-sense (SS). While Cas9 was under constitutive expression, repeat-spacer and tracrRNA were under arabinose inducible promoters. Therefore, the addition of 10 ug/ml of arabinose added during transformation recovery was compared. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | |||
| + | <div> | ||
| + | <center> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:British_Columbia/Project2"> | ||
| + | <img width="180" class="icon" src="https://static.igem.org/mediawiki/2013/7/79/UBCReturnArrow.png" | ||
| + | onmouseover="this.src='https://static.igem.org/mediawiki/2013/7/71/UBCReturnProject.png'" | ||
| + | onmouseout="this.src='https://static.igem.org/mediawiki/2013/7/79/UBCReturnArrow.png'"/></a> | ||
| + | </center> | ||
| + | </div> | ||
| + | |||
| + | <div align="right"><a href="#top">Top of Page</a></div> | ||
| + | </html> | ||
Latest revision as of 02:22, 29 October 2013
iGEM Home



CRISPR
What are CRISPRs and how do they work?
CRISPRs (Clustered Regularly Interspaced Short Palindromic Repeats) are specific regions in some bacterial and archaeal genomes that, together with associated Cas (CRISPR-associated) genes, function as an adaptive immune system in prokaryotes. While the specific ‘adaptive’ nature of this immunity is still under investigation, it is known that exogenous DNA is processed by Cas proteins into short (~30 base pair) sequences that are adjacent to the Protospacer Adjacent Motif (PAM) site. These short pieces of DNA are then incorporated into the host genome between repeat sequences to form spacer elements. The repeat-spacer-repeat array is constitutively expressed (pre-CRISPR RNAs or pre-crRNAs) and processed by Cas proteins to form small RNAs (crRNAs). The small RNAs are then loaded into Cas proteins and act to guide them to initiate the sequence-specific cleavage of the target sequence.
What did we do with them?
As an adaptive immune response, we wanted to know if the CRISPR system could be put together in a modular way to confer resistance to known phage genomes, thereby vaccinating the host. We first wrote scripts capable of identifying the most broadly neutralizing spacer regions from compiled phage genomes. We then assembled the minimum CRISPR components into BioBricks and conducted the necessary proof-of-concept experiments in E. coli. First, we characterized the dynamics of phage infection in our specific host strain and experimental protocols. We then built a system that protects E.coli against T4 phage infection and are beginning to understand some guidelines for assembling CRISPR components to provide immunity. Currently, we are carrying out further validation experiments, extending our system to work with T7 phage, performing some in vitro characterizations, and exploring new possibilities with our working and the tunable CRISPR system.
Why do this?
Ultimately, we hope that large-scale fermenters could be vaccinated to prevent bacterial population collapse caused by environmental phage infection. To extend the application of this approach, we designed specific neutralization elements that allow for population level programming of a culture. We envisioned this first being applied to yogurt where the biosynthesis of flavour combinations might be controlled by targeted phage addition. Check out our population control section for further details on the construction of a few of these pathways. Approaching this from another angle, our modeling section aims to describe the population dynamics of both vulnerable and immunized populations under phage predation with the hopes of eventually being able to rationally tune relative proportions of flavour compounds.
System Design
The three stages of CRISPR immunity (spacer acquisition, crRNA biogenesis, and target interferences), and the general gene architecture described above, are hallmarks of the CRISPR/Cas system. There is, however, substantial diversity in the naturally occurring spacer elements, the Cas genes and the functional PAM sequences. Approximately 45 different gene families have been identified, which, when combined with analysis of their repeat and PAM sequences, can lead to their categorization into three main groups (Types I, II and III) with 10 subtypes (IA, IB, IC, ID, IE, IF, IIA, IIB, IIIA and IIIB).
We chose to work with the type IIA CRISPR3 locus from Streptococcus thermophilus because it has been previously shown to confer immunity to double stranded DNA (dsDNA) phage and plasmids in Escherichia coli and it requires a minimum set of only three components—one protein (Cas9), a non-coding small RNA (trans-activating crRNA, tracrRNA), and the repeat-spacer array. These three necessary components were decoupled from their native regulatory framework and assembled.

Figure 1. SDS-PAGE of Cas9 in 1% arabinose induction.
Cas9
Cas9 is the hallmark of the type II system—a massive 163 kDa multifunctional protein involved in the processing of pre-crRNA and the cleavage of exogenous DNA. The Cas9 gene was subcloned out of [http://www.addgene.org/39314/ pMJ824] and assembled under both the arabinose inducible pBAD [http://parts.igem.org/Part:BBa_I13453 BBa_I13453] and constitutive [http://parts.igem.org/Part:BBa_J23118 BBa_J23118] promoters. Cas9 was submitted to the registry as part [http://parts.igem.org/Part:BBa_K1129006 BBa_K1129006]. Expression of Cas9 under both arabinose induction and constitutive expression was confirmed with SDS-PAGE.
Trans-activating crRNA
The tracrRNA is a 36-base, small, non-coding RNA sequence of which 25 bases is complementary to the repeat sequence. The hybridization of the tracrRNA to the repeat sequence in the pre-crRNA results in double-stranded RNA that is recognized, and cleaved by, a non-Cas RNaseIII enzyme. It has also been shown that tracrRNA, the mature crRNA and Cas9 are all required for cleavage of target dsDNA.
The tracrRNA was cloned from annealed oligonucleotides and assembled under the arabinose inducible pBAD [http://parts.igem.org/Part:BBa_I13453 BBa_I13453] promoter. It was submitted to the registry as parts [http://parts.igem.org/Part:BBa_K1129008 BBa_K1129008] (sense) and [http://parts.igem.org/Part:BBa_K1129009 BBa_K1129009] (anti-sense).
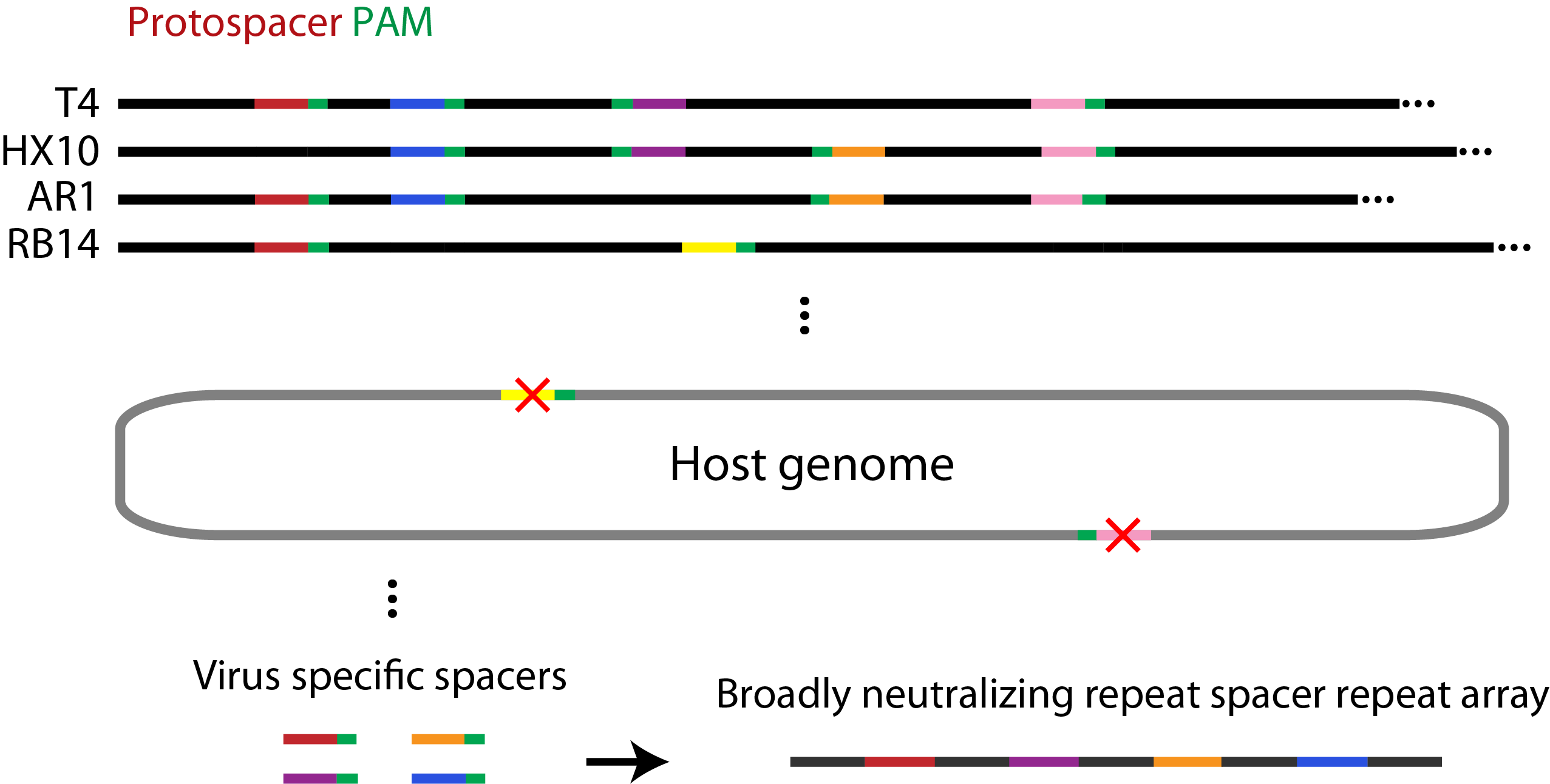
Spacer Selection
Spacers were selected using a perl script (example here) which searches a set of viral genomes to find any occurrences of the PAM site NGGNG. Spacer sequences were removed from the set if (1) the 9 bases preceding the PAM were also found adjacent to a PAM site in the host genome, or (2) if they contained illegal restriction sites. For the remaining spacers, the 30 bases prior to the PAM site were recorded and their frequency of occurrence in each genome was determined.
Figure 2. Summary of the bioinformatics approach used to identify spacer elements broadly neutralizing to several species of phage.
We reasoned the spacers that are the most heavily conserved among related genomes would be under the greatest selective pressure to be retained and would provide a wider range of immunity. Based on this, we selected the spacers which were the most widely distributed in the T7, T4, and closely related genomes for our system. By calculating the fraction of the phage genomes that would be neutralized by combining the most widely distributed spacers, we showed that using our approach only 5 and 3 spacers would be required to neutralize all of the phage related to T4 and T7, respectively.
Toxic Plasmids

Figure 3. Toxic plasmids. CRISPR-mediated cleavage of these plasmids will result in removal of ampicillin resistance, and cause cell death on selective media.
In order to assay for CRISPR-mediated defence against plasmids, we designed two plasmids that, when cleaved, will eliminate the antibiotic resistance conferred by the “invading” plasmid, thereby resulting in cell death. These two plasmids contain the spacer and PAM sequence identified by our informatics pipeline and an ampicillin resistance marker. They have been submitted to the registry (with chloramphenicol resistance) in parts [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1129010 BBa_K1129010] (T4) and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1129011 BBa_K1129011] (T7).
Results
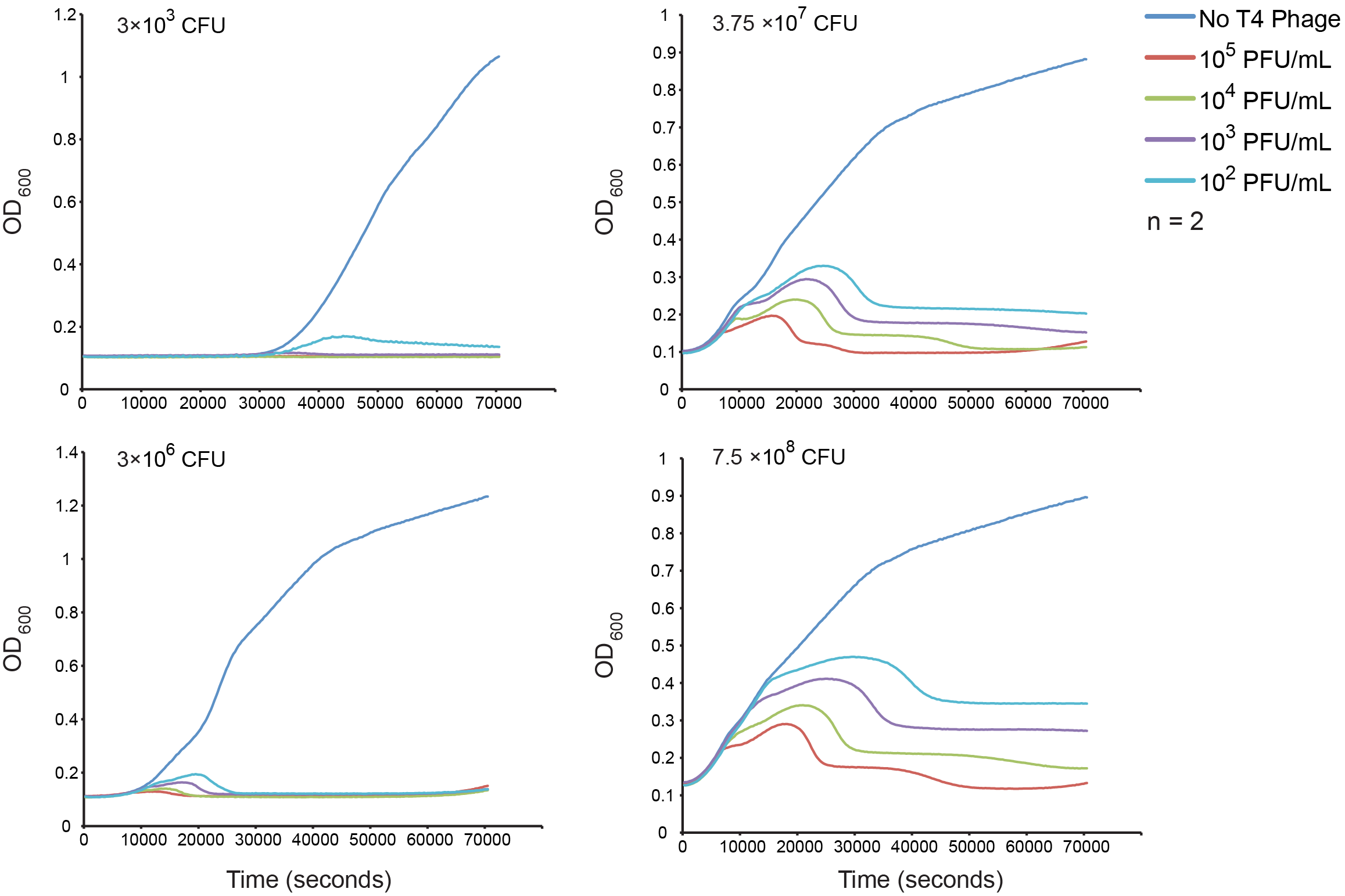
Normal growth under T4 Phage Infection
Our first goal was to measure bacterial growth kinetics under T4 phage predation with our host strain and experimental conditions. The upper left quadrant has the estimated inoculums and four different dilutions of phage were assessed. The results are consistent with a step-wise lysis cycle that is well documented in the literature. See the modeling section for more detailed analysis of the kinetics.
Figure 4. The effect of initial phage concentration on E. coli GB10 cell growth for various inoculums. Overnight cultures of wild-type E. coli 10G cells were grown at 37 °C until an OD of 0.3 was obtained diluted to each inoculum, the cells counts of which were estimate by plate counts (CFU). The cultures were grown in a 96-well format and OD was measured every 5 min for 2 hrs (n = 2).
Combinatorial CRISPR-Cas9 element assembly
Once we were confident in our ability to measure growth under phage predation, we designed a strategy to assemble Cas9, tracrRNA (sense/antisense), and the T4 spacer-repeat-spacer with inducible promoters in a combinatorial pattern and made a library of approximately 800 clones (see protocols). We then added 103 phage to each clone and screened for growth in high-throughput.
Figure 5. Measuring OD of combinatorial library clones after incubation for 24 hours at 30 °C.The horizontal red line represents the threshold for selection of clones for sequencing and colored points correspond to characterization data in the subsequent figure. 103 phage were added to each well.
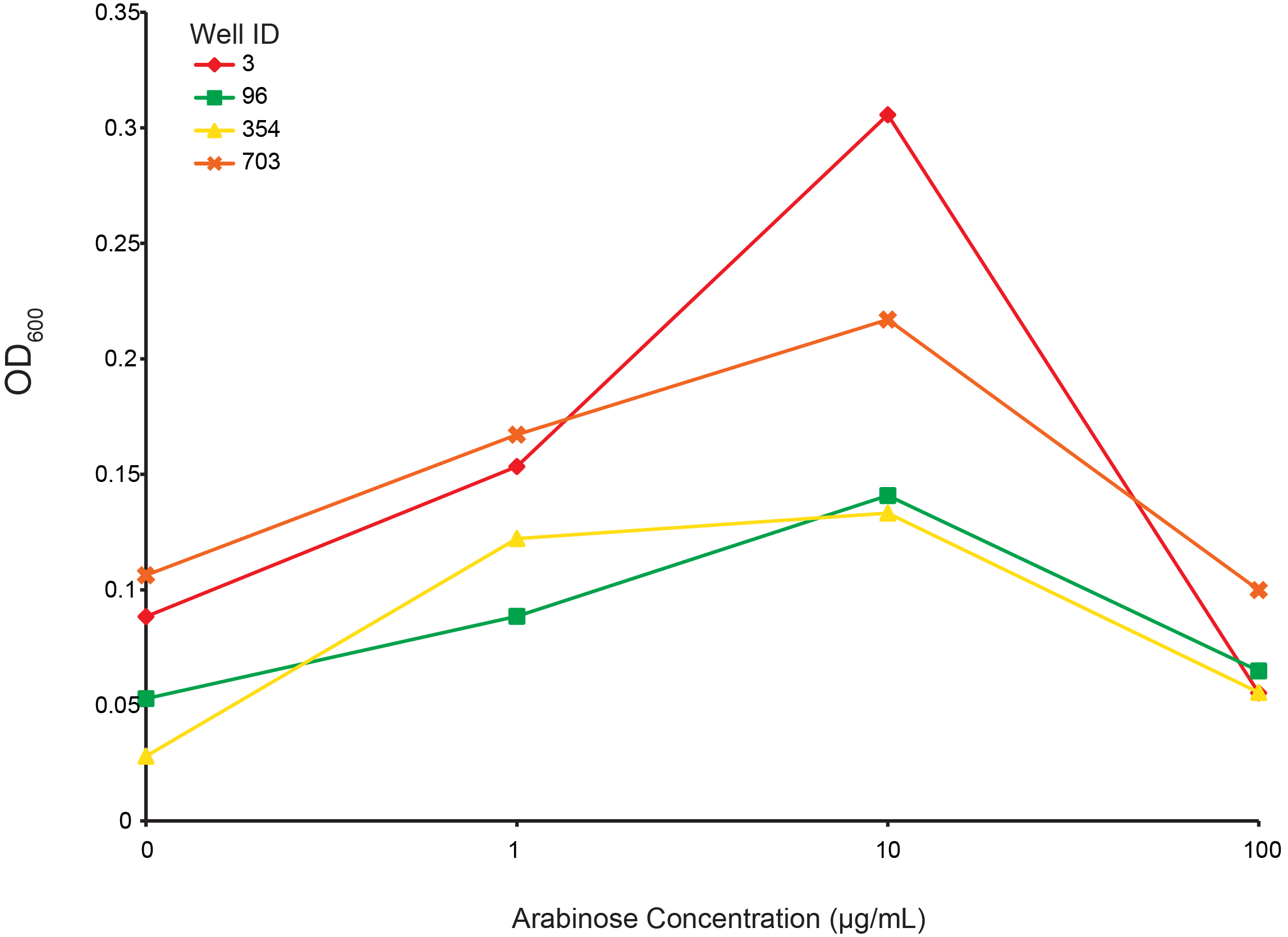
Arabinose dependent growth under T4 infection
From the preceding experiment, we selected four clones for characterization (highlighted in figure 2; red, green, yellow, orange are wells 3, 96, 354, 703, respectively). As all the CRISPR components were on arabinose inducible promoters, we looked for the arabinose dependant survival of selected clones under phage predation. An intermediate level of induction gave the largest final OD and suggests that high induction of Cas9 or other components creates burdens on the cell. These data also supports the CRISPR construct being responsible for escaping phage predation.
Figure 6. Measuring final OD of select combinatorial library clones under different arabinose concentrations after incubation for 24 hours at 30 °C. Well IDs correspond with the precending figure. 103 phage were added to each well.
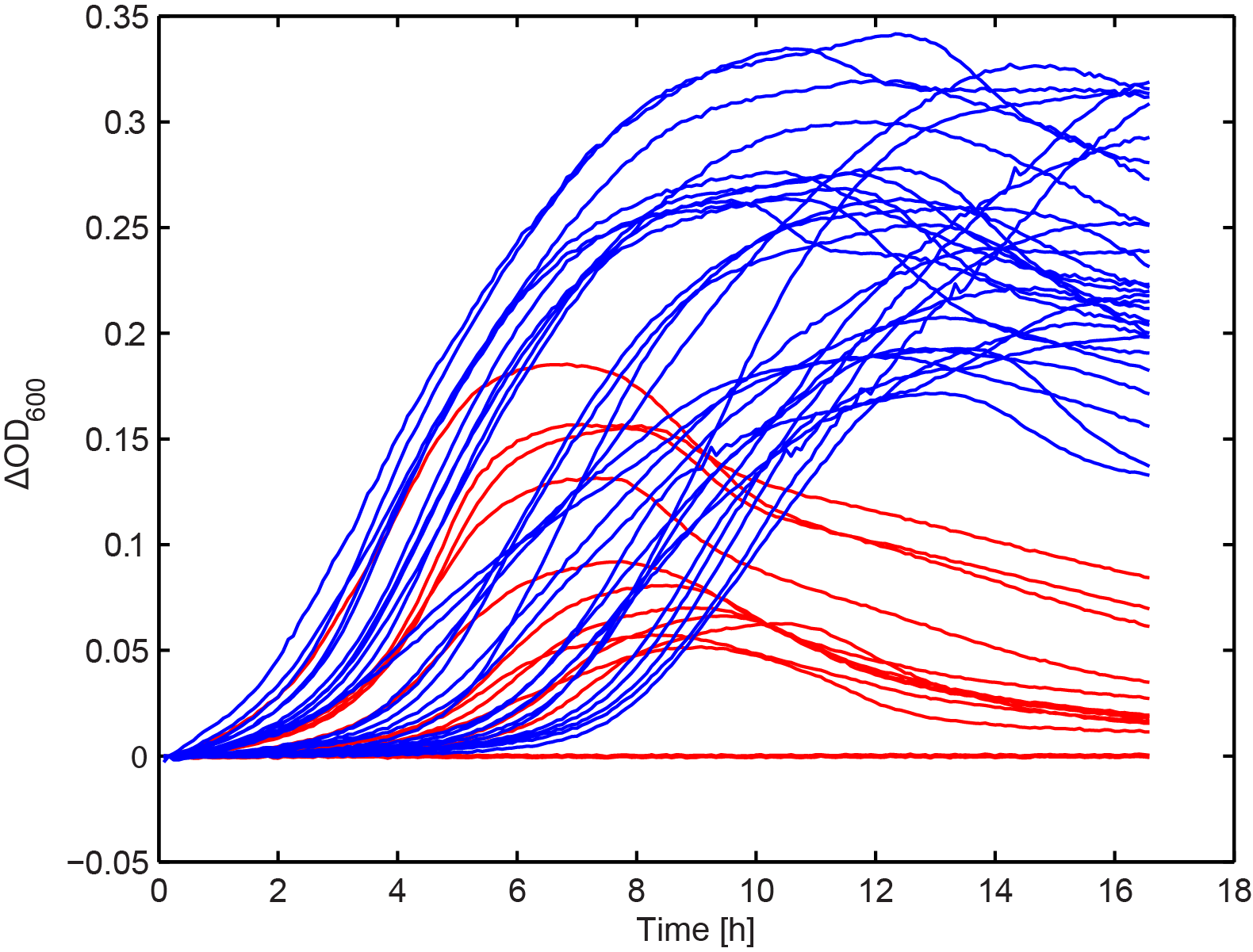
Growth kinetics of immunized culture
From the combinatorial library, we performed colony PCR on 30 clones and showed that the majority of constructs had an approximately 5 kb long insert. A subset of these, along with the clones selected in Figure 2, were arrayed into a 96-well plate. We then added 103 T4 phage to each well and measured OD in the same way as in Figure 1. This seemed to show that there were two distinct groups, one with increased fitness in the presence of phage.
Figure 7. The effect of phage predation on clones from combinatorial libraries. Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 103 phage were then added to each well and OD was monitored overnight. The results seem to resolve two groups, one showing increased final OD over the other (blue and red lines respectively) (n=2).
Finally, we added a inoculum of cells that corresponded our initial characterization of phage predation (Figure 1) and monitored growth of two select clones from the two different groups identified in the preceding experiment (Figure 4). This showed an interesting dip in growth at approximately 10,000 seconds, which we are in the process of characterizing. Also, we have yet to confirm the exact orientation and composition of the clones from the combinatorial library. Once this is done, however, we are expecting to be able to comment on the minimum set of CRISPR components required for resistance and on any difference with sense vs. antisense tracrRNA.
Figure 8. Monitoring growth of two select combinatorial library clones under high cell inoculum. 103 phage were then added to each well and OD was monitored overnight (n=2).
Testing host background dependance
To ensure that the survival phenotype we were observing was in-fact dependant on the expected constructs, we re-isolated plasmid from "survivors" and re-assayed their growth under phage predation. We also sent these plasmids for sequencing as to assign a genotype that resulted from our combinatorial assembly. In a new host background, the growth advantage was not maintained, suggesting that the phenotype was dependant on changes occurring in the host background (Figure 9). Furthermore, the plasmid sequences showed that the repeat-spacer regions was not present, showing that the combinatorial screening strategy did now work as expected. There could, however, still be something interesting happening where the host strain is able to acquire resistance more readily while expressing Cas9. There was also only a slight correlation between the presence of tracrRNA and survival. We are looking at this more closely. More importantly, we need to be careful looking using high-throughput approaches as this increases the rate at which spontaneous processes can confound the results.
Figure 9. The effect of phage predation on clones from combinatorial libraries after re-transformation and genotyping. Change in OD in culture in a 96-well plate grown for 11 hours at 30 °C. 103 phage were then added to each well and OD was monitored overnight (n=3). Clones containing either tracrRNA or spacers are shown in green.
Testing the standard assembled construct
We then finished the standard assembly of our designed CRISPR construct and tested growth under T4 phage predation. The construct contained cas9, the repeat-spacer region and both orientations of the tracrRNA. This showed an interesting phenotype where the entire construct seems to cause enhanced recovery after a cycle of phage predation (starting at approximately 25 hrs). The initial infection cycle (from 0 to 15 hrs) showed that cas9 caused a significant hindrance in growth compared to an empty vector and the repeat-spacer/tracrRNA control (Figure 9).
Figure 9. The effect of phage predation on clones from assembled CRISPR constructs all under inducible promoters: cas9/repeat-spacer/tracrRNA-sense (AAS), repeat-spacer/tracrRNA-sense (SS), and empty vector (control). Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 103 phage and 1 ug/ml of arabinose was then added to each well and OD was monitored for 46 hrs (n=2).
We then monitored this trend by taking final OD after 46 hrs and ensured that the phenotype was consistent under various arabinose and phage concentrations. Figure 10 shows final OD with a higher phage inoculum.
Figure 10. The effect of phage predation on clones from assembled CRISPR constructs all under inducible promoters: cas9/repeat-spacer/tracrRNA-sense (ASS), repeat-spacer/tracrRNA-sense (SS), and empty vector (control). Clones were arrayed into a 96-well plate and grown for 6 hours at 30 °C. 104 phage and 1 ug/ml of arabinose was then OD was measured after 46 hrs.
Transformation assay
While the phage phenotypes show promise for providing a growth advantage under phage, we also wanted to show some evidence for the sequence specificity of our CRISPR construct. The experimental approach is also described above in the toxic plasmids section. We designed a transformation assay which mixed a RFP containing control plasmid and a decoy plasmid which contained the targeted T4 and PAM sequence (both on PSB1A3, to be maintained along with PSB1C3 under both ampicillin and chloramphenicol selection). We would then expect that if the decoy plasmid was being cleaved, the ratio of red/white colonies counted on a plated would increase as the decoy plasmid was being cleaved. This was in-fact observed for the entire construct, and controlled for using a construct that was missing Cas9 (Figure 11).
Figure 11. Colony counts of RFP and decoy plasmid containing clones (red and white, respectively): Cas9/repeat-spacer/tracrRNA-sense (CSS) and repeat-spacer/tracrRNA-sense (SS). While Cas9 was under constitutive expression, repeat-spacer and tracrRNA were under arabinose inducible promoters. Therefore, the addition of 10 ug/ml of arabinose added during transformation recovery was compared.
 "
"