Team:Linkoping Sweden/Notebook
From 2013.igem.org
m |
|||
| (14 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
<script type="text/javascript"> | <script type="text/javascript"> | ||
| - | var page= | + | var page=5; |
$("#cfcontrols li").eq(page).addClass("active"); | $("#cfcontrols li").eq(page).addClass("active"); | ||
$("#cfcontrols li a span").eq(page).addClass("selected"); | $("#cfcontrols li a span").eq(page).addClass("selected"); | ||
| Line 8: | Line 8: | ||
</script> | </script> | ||
</html> | </html> | ||
| + | |||
| + | ---- | ||
'''Construction of a living detection system or biosensor of allergens using Luciferase''' | '''Construction of a living detection system or biosensor of allergens using Luciferase''' | ||
| - | A common problem in todays society is allergies. To be able to quickly detect the | + | A common problem in todays society is allergies. To be able to quickly detect the presence of an antigen we are developing a biosensor. The sensor is created by letting genetically modified E.Coli produce a complex consisting of an antibody and a fluorescent protein. When this complex comes in contact with a certain antigen attached to another fluorescent protein, it will emit light. This light will be discovered by a detector and translated into an electrical signal. Using this method, very small amounts of an antigen can be measured and amplified. |
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | '''September 11''' | ||
| + | |||
| + | Great success today as we managed to create our first biobrick! The band in the red circle on the gel shows us that we got the antibody, promoter and luciferase together to create a new biobrick. The next step will be to insert this biobrick into our bacteria. | ||
| + | |||
| + | [[File:Gele biobrick AK.png]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''August 17''' | ||
| + | |||
| + | After adding enzymes to cut our Luciferase part from the plasmid, we did a gel electrophoresis. In this way, we can control that the enzymes cut at the right places in the plasmid, resulting in a fragment about 3000 base pairs long for Luciferase. After that we just take the Luciferase band visible on the gel, cut it out, and smash the gel to prepare it for further purification steps. | ||
| + | |||
| + | [[File:linnea_labbar.png]] | ||
| + | [[File:linnea_anna.png]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''July 24''' | ||
| + | |||
| + | Today we were interviewed on the Swedish radio channel ”Sveriges Radio P4 Östergötland” | ||
| + | Anna and David got to explain what we are trying to do in our project and inform people about synthetic biology and the iGEM competition. They did a great job! If you want to listen to the interview (in Swedish) just click on the link below! | ||
| + | |||
| + | http://sverigesradio.se/sida/artikel.aspx?programid=2949&artikel=5600104 | ||
| + | |||
| + | [[File:radio_julle_anna.png]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''July 23''' | ||
| + | |||
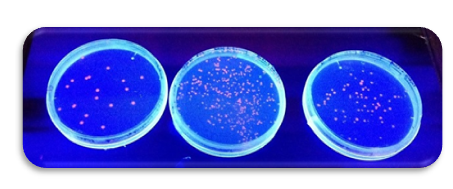
| + | After some attempts we finally got a good transformation! The red dots glowing in the UV-light are E.Coli that during the electroporation successfully managed to take up the DNA for RFP (Red Fluorescent Protein) and started to produce this. | ||
| + | |||
| + | [[File:plattor.png]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''June 3''' | ||
| + | |||
| + | To start with, we needed to find some E.Coli. With the help from Cecilia at LiU, we got a compatible stem of E.Coli, ready for electroporation. To assure that all concentrations were right, that the agar was correctly prepared, and that the bacteria would survive the electroporation and grow as desired, we decided to do some transformation attempts using only RFP and Luciferase. By doing this, we increase our chances of getting the best possible result with the Antibody-Complex DNA later on. | ||
| + | |||
| + | [[File:labb.png]] | ||
| + | |||
| + | {{Template:Team Linkoping footer}} | ||
Latest revision as of 09:50, 21 October 2013














Construction of a living detection system or biosensor of allergens using Luciferase
A common problem in todays society is allergies. To be able to quickly detect the presence of an antigen we are developing a biosensor. The sensor is created by letting genetically modified E.Coli produce a complex consisting of an antibody and a fluorescent protein. When this complex comes in contact with a certain antigen attached to another fluorescent protein, it will emit light. This light will be discovered by a detector and translated into an electrical signal. Using this method, very small amounts of an antigen can be measured and amplified.
September 11
Great success today as we managed to create our first biobrick! The band in the red circle on the gel shows us that we got the antibody, promoter and luciferase together to create a new biobrick. The next step will be to insert this biobrick into our bacteria.
August 17
After adding enzymes to cut our Luciferase part from the plasmid, we did a gel electrophoresis. In this way, we can control that the enzymes cut at the right places in the plasmid, resulting in a fragment about 3000 base pairs long for Luciferase. After that we just take the Luciferase band visible on the gel, cut it out, and smash the gel to prepare it for further purification steps.
July 24
Today we were interviewed on the Swedish radio channel ”Sveriges Radio P4 Östergötland” Anna and David got to explain what we are trying to do in our project and inform people about synthetic biology and the iGEM competition. They did a great job! If you want to listen to the interview (in Swedish) just click on the link below!
http://sverigesradio.se/sida/artikel.aspx?programid=2949&artikel=5600104
July 23
After some attempts we finally got a good transformation! The red dots glowing in the UV-light are E.Coli that during the electroporation successfully managed to take up the DNA for RFP (Red Fluorescent Protein) and started to produce this.
June 3
To start with, we needed to find some E.Coli. With the help from Cecilia at LiU, we got a compatible stem of E.Coli, ready for electroporation. To assure that all concentrations were right, that the agar was correctly prepared, and that the bacteria would survive the electroporation and grow as desired, we decided to do some transformation attempts using only RFP and Luciferase. By doing this, we increase our chances of getting the best possible result with the Antibody-Complex DNA later on.






 "
"