Team:Paris Saclay
From 2013.igem.org
(Prototype team page) |
|||
| (35 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Team:Paris_Saclay/incl_debut_generique}} | |
| + | = '''Welcome to the Paris-Saclay iGEM 2013 Wiki ! ''' = | ||
| - | + | <div style="border:1px solid #DEDEDE;background-color:#DEDEFF;padding-left:3px;">'''Note : '''this wiki uses modern web standards. Please use a recent browser (tested on Firefox 16+ & Chrome 27+).</div> | |
| - | <div | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </div | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == Our project : The PCB Busters !== | |
| + | [[File:Pshome.jpg|right|300px|caption]] | ||
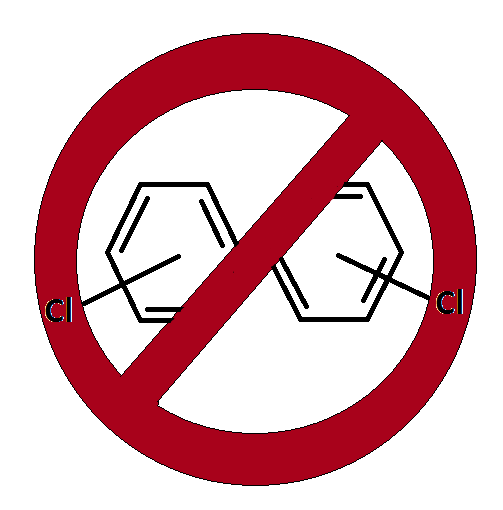
| + | [[Team:Paris_Saclay/PCBs|PCBs (Polychlorobiphenyls)]] are synthetic chemicals that were widely used in electrical equipment during the late 20th century. These hazardous organic compounds are extraordinary stable and not readily biodegradable. Thus they remain for many years in nature and belong to family of persistent pollutants. PCBs accumulate in animal fatty tissues due to their lipophilicity, starting from aquatic organisms and going up the food chain. Indeed, they can be found in high concentrations in human adipose tissues. As most PCBs are probably carcinogenic (class 2A) and some are recognized endocrine disruptors, they may lead to important health issues. This is especially true in France where a study showed higher levels of PCB in French people adipose tissues than in adipose tissues of people from other parts of the world. | ||
| - | + | Although PCBs are man-made and have no natural equivalents, some bacterial communities have evolved and developed the capacity to degrade PCBs. Degradation of highly chlorinated PCBs begins with an anaerobic reductive dechlorination that lowers the number of chlorine atoms bear by the PCB molecules. PCBs with few chlorine atoms can then be degraded via the biphenyl degradation pathway. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:Pshome1.png|left|100px|caption]] | |
| - | + | Our project aims at '''<span style="color:#E60000">constructing an ''Escherichia coli'' strain capable of degrading highly chlorinated PCBs</span>'''. This will be achieved by introducing in an ''E. coli'' strain genes involved in the degradation of PCBs from the three most efficient PCB degrading strains described so far: ''Pseudomonas pseudoalcaligenes'', ''Burkholderia Xenovorans'' and ''Rhodoccocus jostii''. Because the first degradation steps are anaerobic and later steps are aerobic, we want to '''<span style="color:#E60000">introduce an oxygen based regulation of the expression of the different genes</span>''' involved in both aerobic and anaerobic degradation steps. Finally, we are also '''<span style="color:#E60000">developing a sensor system to detect PCBs</span>''' in the environment. | |
| - | + | ||
| - | + | [[File:Psphoto.jpg|center|400px|caption]] | |
| - | + | ||
| - | + | ||
| - | + | As part of our project, we had a lot of [[Team:Paris_Saclay/Human_Practices|reflection]] about the safety issues that our project should overcome, not only between the team members, but also with researchers of our Institute and non scientific people. | |
| - | + | We also decided to explore the [[Team:Paris_Saclay/Open_source|Open Source]] concepts at the heart of the iGEM competition. This is why we organised and attended some events and interviews related to Open Source biology and the implications of DIY synthetic biology. We also decided to build our own [https://2013.igem.org/Team:Paris_Saclay/PS-PCR PCR thermal cycler]. | |
| - | + | ||
| - | + | {{Team:Paris_Saclay/incl_fin}} | |
| - | + | ||
| - | + | ||
Latest revision as of 17:48, 4 October 2013
Welcome to the Paris-Saclay iGEM 2013 Wiki !
Our project : The PCB Busters !
PCBs (Polychlorobiphenyls) are synthetic chemicals that were widely used in electrical equipment during the late 20th century. These hazardous organic compounds are extraordinary stable and not readily biodegradable. Thus they remain for many years in nature and belong to family of persistent pollutants. PCBs accumulate in animal fatty tissues due to their lipophilicity, starting from aquatic organisms and going up the food chain. Indeed, they can be found in high concentrations in human adipose tissues. As most PCBs are probably carcinogenic (class 2A) and some are recognized endocrine disruptors, they may lead to important health issues. This is especially true in France where a study showed higher levels of PCB in French people adipose tissues than in adipose tissues of people from other parts of the world.
Although PCBs are man-made and have no natural equivalents, some bacterial communities have evolved and developed the capacity to degrade PCBs. Degradation of highly chlorinated PCBs begins with an anaerobic reductive dechlorination that lowers the number of chlorine atoms bear by the PCB molecules. PCBs with few chlorine atoms can then be degraded via the biphenyl degradation pathway.
Our project aims at constructing an Escherichia coli strain capable of degrading highly chlorinated PCBs. This will be achieved by introducing in an E. coli strain genes involved in the degradation of PCBs from the three most efficient PCB degrading strains described so far: Pseudomonas pseudoalcaligenes, Burkholderia Xenovorans and Rhodoccocus jostii. Because the first degradation steps are anaerobic and later steps are aerobic, we want to introduce an oxygen based regulation of the expression of the different genes involved in both aerobic and anaerobic degradation steps. Finally, we are also developing a sensor system to detect PCBs in the environment.
As part of our project, we had a lot of reflection about the safety issues that our project should overcome, not only between the team members, but also with researchers of our Institute and non scientific people.
We also decided to explore the Open Source concepts at the heart of the iGEM competition. This is why we organised and attended some events and interviews related to Open Source biology and the implications of DIY synthetic biology. We also decided to build our own PCR thermal cycler.
 "
"