Team:TU-Delft/Project
From 2013.igem.org
| (66 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:TU-Delft/Templates/Navigation}} | {{:Team:TU-Delft/Templates/Navigation}} | ||
{{:Team:TU-Delft/Templates/Style}} | {{:Team:TU-Delft/Templates/Style}} | ||
| + | {{:Team:TU-Delft/Templates/Frog}} | ||
| + | {{:Team:TU-Delft/Templates/Logo}} | ||
| + | |||
<html> | <html> | ||
| - | < | + | <div style="margin-left:30px;margin-right:30px;margin-top:0px; width:900px;float:left;"> |
| + | <center> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/8/82/Pep9.jpg"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/8/82/Pep9.jpg" alt="" HEIGHT="90" WIDTH="700" /> | ||
| + | </a> | ||
| + | </center> | ||
<br> | <br> | ||
| + | <p align="justify"> | ||
| + | The bacterium Methicillin-Resistant <i>Staphylococcus aureus</i> causes major problems, especially in hospitals, causing over half a million infections annually in the United States alone. Of the alternative treatments currently under investigation one of the more promising is antimicrobial peptides (AMPs). These small peptides, typically several dozen amino acids in length, bind to and aggregate on the outer-membrane of the target organism and are able to make holes in the membrane at sufficiently high concentrations. The variation in the membrane composition of different organisms makes these peptides highly specific, meaning that infectious bacteria can be killed with little effect on neighboring human cells. Thousands of AMPs are have been discovered in nature and unlike antibiotic chemicals, AMPs can easily evolve as pathogens develop resistance. Using multiple AMPs in combination, can reduce the probability of developing resistance. The Peptidor project consists of an <i>E. coli</i> that can detect S. aureus in order to locally produce and deliver AMPs at the site of infection. Detection is achieved using the native quorum sensing system of S. aureus. Upon detection, AMPs, inactivated by fusion to a SUMO-tag, are overexpressed. After a delay period, introduced through a negative transcriptional cascade, a SUMO protease is expressed which cleaves off the inactivating tag of the peptides. Additionally the timer activates a safety mechanism containing an <i>E. coli</i> kill switch. Using this mechanism, high concentrations of peptide can be delivered very locally in order to efficiently kill <i>S. aureus</i>. </p> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <br> | ||
| + | <center> | ||
| + | |||
| + | <a href="https://static.igem.org/mediawiki/2013/5/51/Schem-new.jpg" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/5/51/Schem-new.jpg" width=650px height=460px> | ||
| + | </a> | ||
| + | </center> | ||
| + | <center> | ||
| + | <p>Figure 1: Schematic diagram of the complete system</p> | ||
| + | </center> | ||
| + | <br> | ||
| + | The project is split up into different modules. Here an overview is given:<br><br> | ||
| + | |||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Sensing" style="text-decoration: none"" target="_blank"><font color="#0080FF" size="3">Sensing device</font></a>: The receiving part of the quorum sensing system of <i>S. aureus</i>, AgrA/AgrC operon, is cloned into our Peptidor in order to detect the presence of <i>S. aureus</i>. | ||
| + | <br><br> | ||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Peptides" style="text-decoration: none"" target="_blank"><font color="#0080FF" size="3">Peptide production</font></a>: After <i>S. aureus</i> has been detected, production of anti-microbial peptide will be started in a manner that allows a strongly localized, high concentration at the exact positition of <i>S. aureus</i>. | ||
| + | </p > | ||
| + | <br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU-Delft/PeptideCharacterization" style="text-decoration: none"" target="_blank"><font color="#0080FF" size="3">Peptide characterization</font></a> The peptides produced are characterized with the use of both SDS-PAGE and MS/MS. | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Timer" style="text-decoration: none"" target="_blank"><font | ||
| + | color="#0080FF" size="3">Timer</font></a>: A timer was added to the circuit in order the peptide pro | ||
| + | |||
| + | for two main reasons. First of all the existence of the timer gives the time for the level of peptide to build up in the cell before activation through cleavage occurs. When activated too early, the expression host could be severally effected or the peptides could get trapped in inclusion bodies. | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Killswitch" style="text-decoration: none"" target="_blank"><font | ||
| + | color="#0080FF" size="3">Kill switch</font></a> :To secure safety for both the patient and the environment, a kill switch is incorporate to ensure lysis of the <i>E. coli</i> expression host. | ||
| + | |||
| + | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
Latest revision as of 18:13, 4 October 2013


The bacterium Methicillin-Resistant Staphylococcus aureus causes major problems, especially in hospitals, causing over half a million infections annually in the United States alone. Of the alternative treatments currently under investigation one of the more promising is antimicrobial peptides (AMPs). These small peptides, typically several dozen amino acids in length, bind to and aggregate on the outer-membrane of the target organism and are able to make holes in the membrane at sufficiently high concentrations. The variation in the membrane composition of different organisms makes these peptides highly specific, meaning that infectious bacteria can be killed with little effect on neighboring human cells. Thousands of AMPs are have been discovered in nature and unlike antibiotic chemicals, AMPs can easily evolve as pathogens develop resistance. Using multiple AMPs in combination, can reduce the probability of developing resistance. The Peptidor project consists of an E. coli that can detect S. aureus in order to locally produce and deliver AMPs at the site of infection. Detection is achieved using the native quorum sensing system of S. aureus. Upon detection, AMPs, inactivated by fusion to a SUMO-tag, are overexpressed. After a delay period, introduced through a negative transcriptional cascade, a SUMO protease is expressed which cleaves off the inactivating tag of the peptides. Additionally the timer activates a safety mechanism containing an E. coli kill switch. Using this mechanism, high concentrations of peptide can be delivered very locally in order to efficiently kill S. aureus.
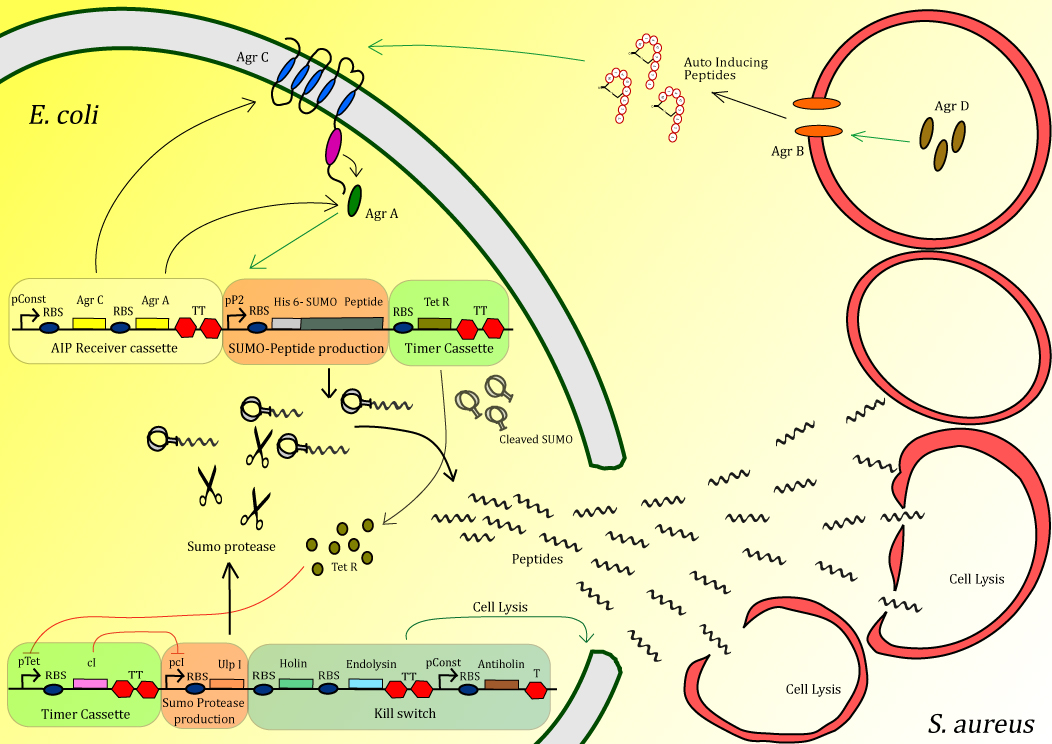
Figure 1: Schematic diagram of the complete system
The project is split up into different modules. Here an overview is given:
Sensing device: The receiving part of the quorum sensing system of S. aureus, AgrA/AgrC operon, is cloned into our Peptidor in order to detect the presence of S. aureus.
Peptide production: After S. aureus has been detected, production of anti-microbial peptide will be started in a manner that allows a strongly localized, high concentration at the exact positition of S. aureus.
Peptide characterization The peptides produced are characterized with the use of both SDS-PAGE and MS/MS.
Timer: A timer was added to the circuit in order the peptide pro for two main reasons. First of all the existence of the timer gives the time for the level of peptide to build up in the cell before activation through cleavage occurs. When activated too early, the expression host could be severally effected or the peptides could get trapped in inclusion bodies.
Kill switch :To secure safety for both the patient and the environment, a kill switch is incorporate to ensure lysis of the E. coli expression host.
 "
"
