Team:Leeds/Project
From 2013.igem.org
(Difference between revisions)
| Line 52: | Line 52: | ||
<br> | <br> | ||
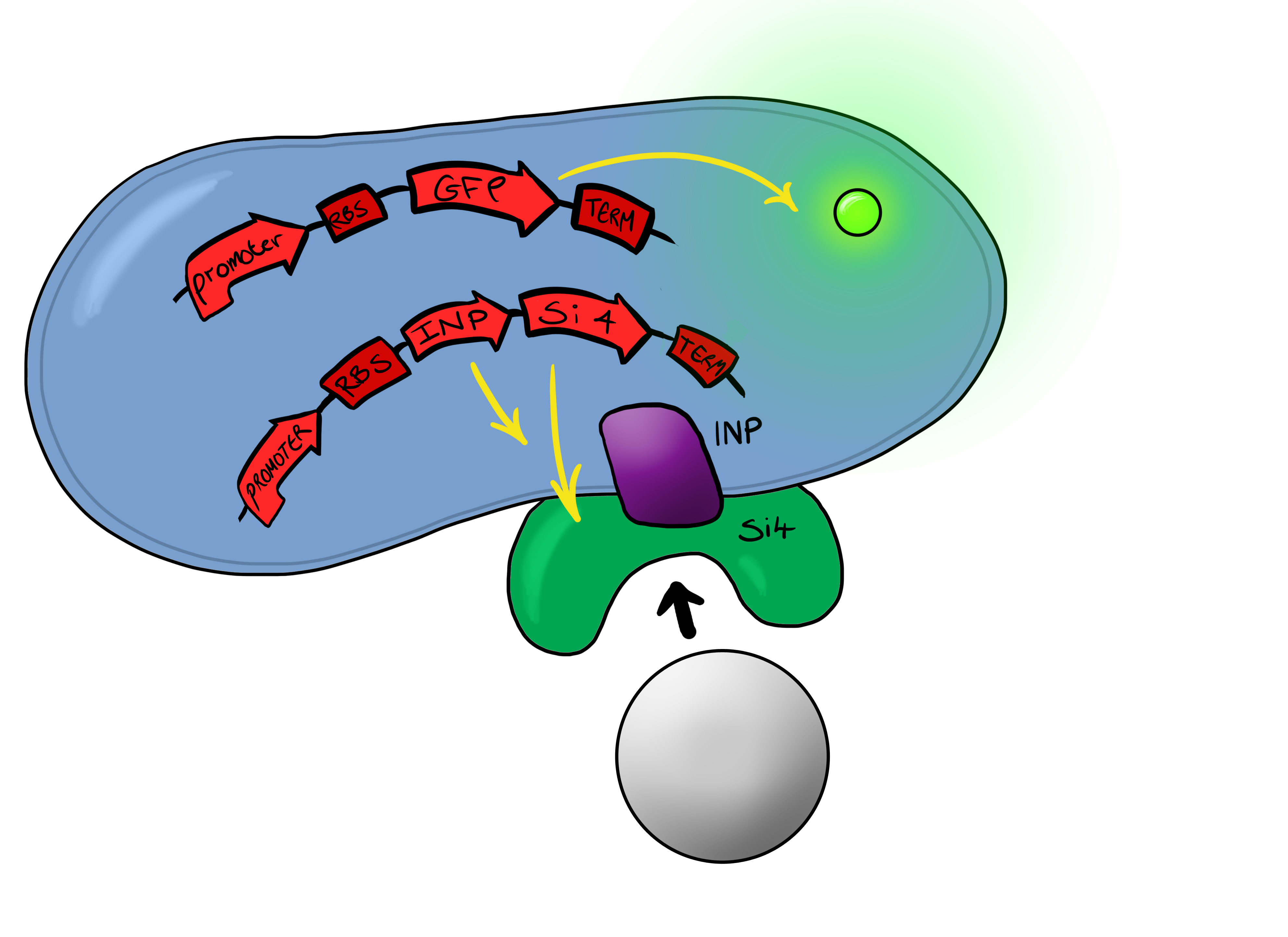
Well, MicroBeagle is a E.coli cell that has been modified so that it can “hunt down” pathogens. By utilising the Cpx pathway and GFP we hope to create a new biosensor that gives results that can be seen with the naked eye. | Well, MicroBeagle is a E.coli cell that has been modified so that it can “hunt down” pathogens. By utilising the Cpx pathway and GFP we hope to create a new biosensor that gives results that can be seen with the naked eye. | ||
| + | <br> | ||
| + | Why is the MircoBeagle a useful device? | ||
| + | <br> | ||
| + | There could be many different possible uses for our device as the pathogen detection mechanism we are using is modular and can easily be tailored to individual needs. We see the MircoBeagle being used for testing blood samples, detection of heavy metals and, our main focus, detection of pathogens in water samples. | ||
| + | <br> | ||
| + | Detecting pathogens in water is a really important application as 3.4 million people a year die from water related diseases. The figures are shocking and we as a team want to create something that may, in the future, help to reduce those numbers. | ||
| + | <br> | ||
| + | Below is a summary of our Biosystem development strategy... | ||
<br> | <br> | ||
==Phase I== | ==Phase I== | ||
| Line 76: | Line 84: | ||
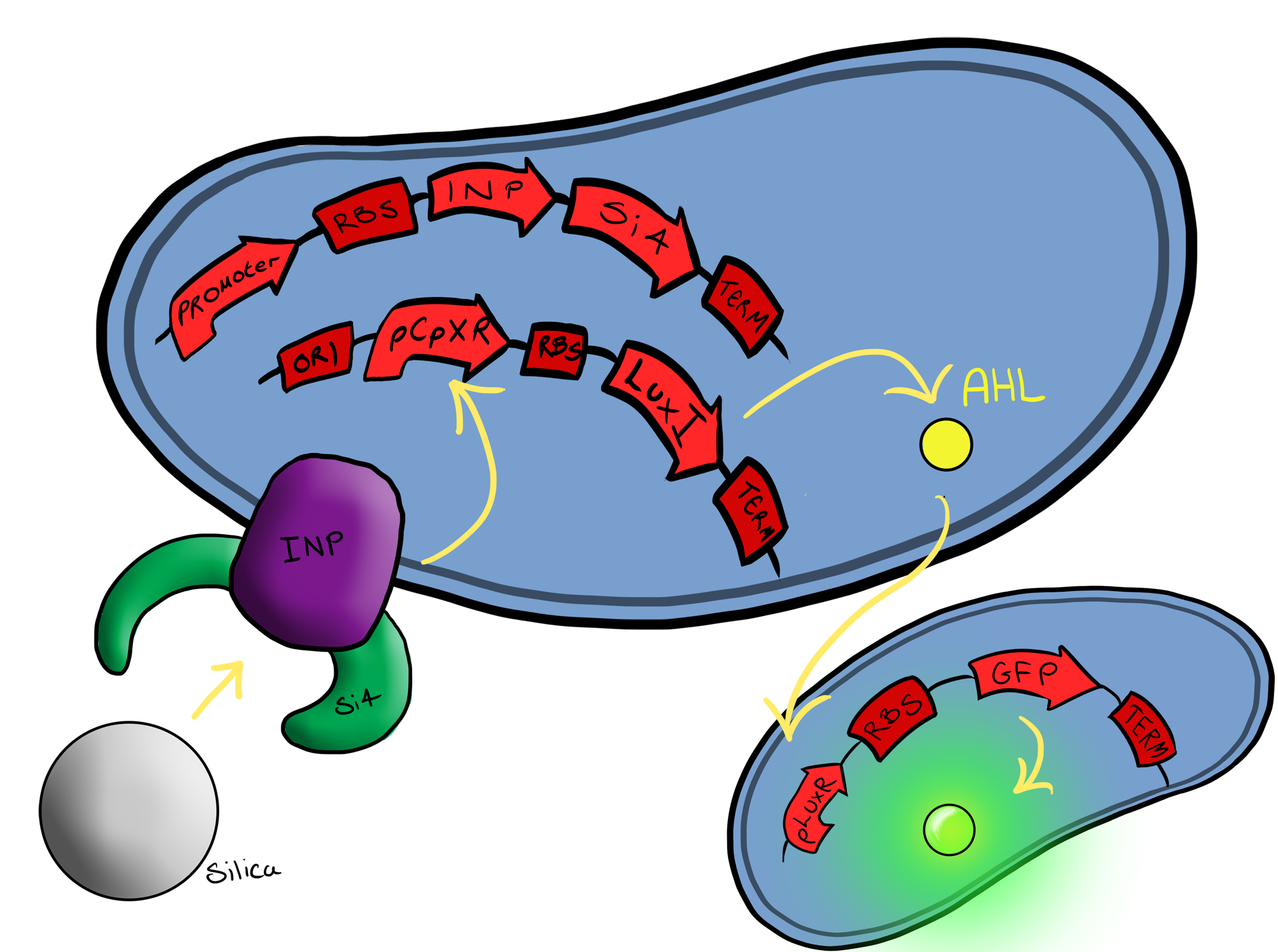
===Biosystem 3=== | ===Biosystem 3=== | ||
[[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | [[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br style="clear:both"/> | <br style="clear:both"/> | ||
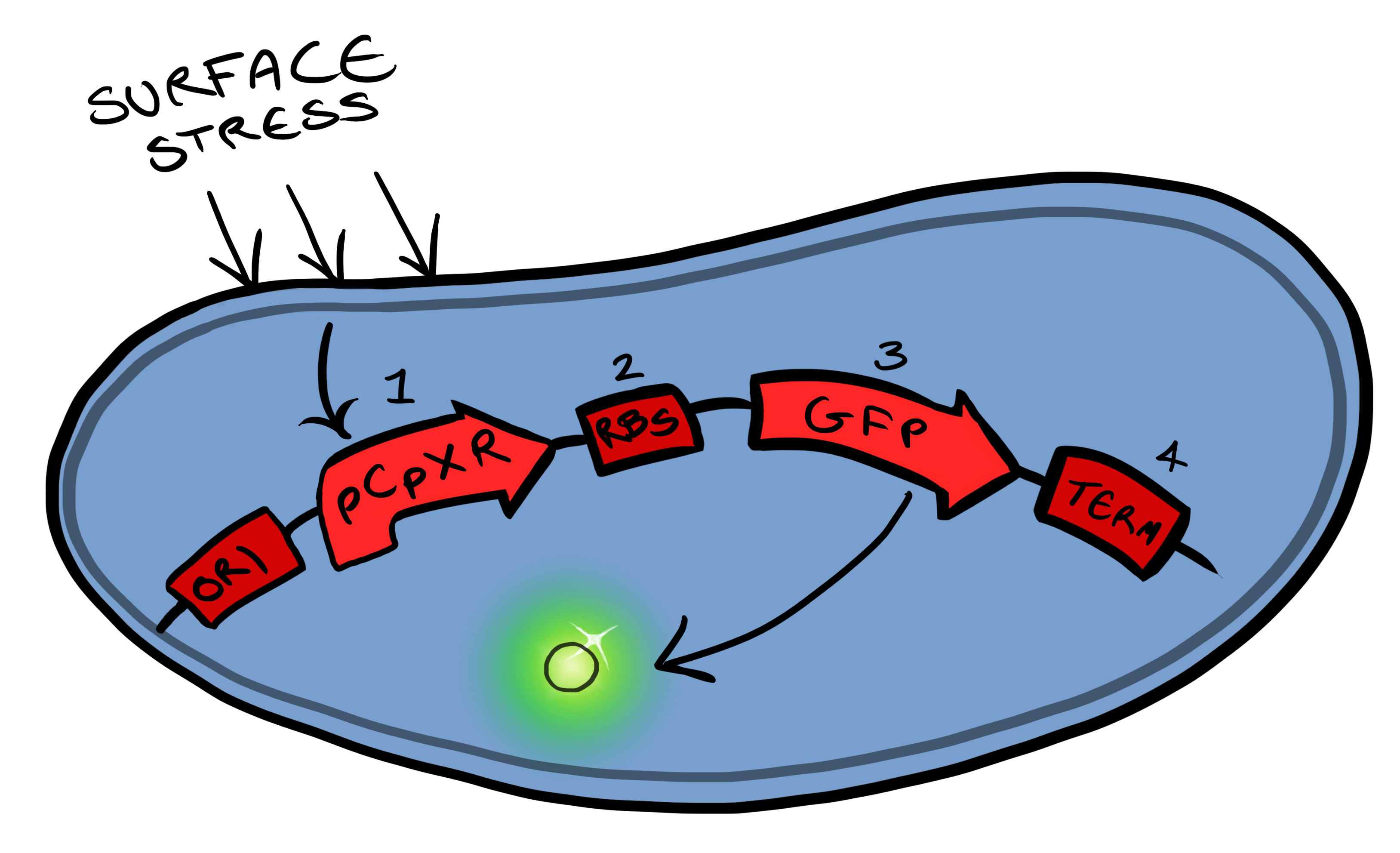
==The Cpx Pathway== | ==The Cpx Pathway== | ||
This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | ||
}} | }} | ||
Revision as of 10:59, 17 September 2013
 "
"