Team:Leeds/Project
From 2013.igem.org
(Difference between revisions)
| Line 71: | Line 71: | ||
===Biosystem 2=== | ===Biosystem 2=== | ||
[[File:Leeds_Device2schematic.png|right|300px|Cartoon schematic of Biosystem 2|link=|frameless]] | [[File:Leeds_Device2schematic.png|right|300px|Cartoon schematic of Biosystem 2|link=|frameless]] | ||
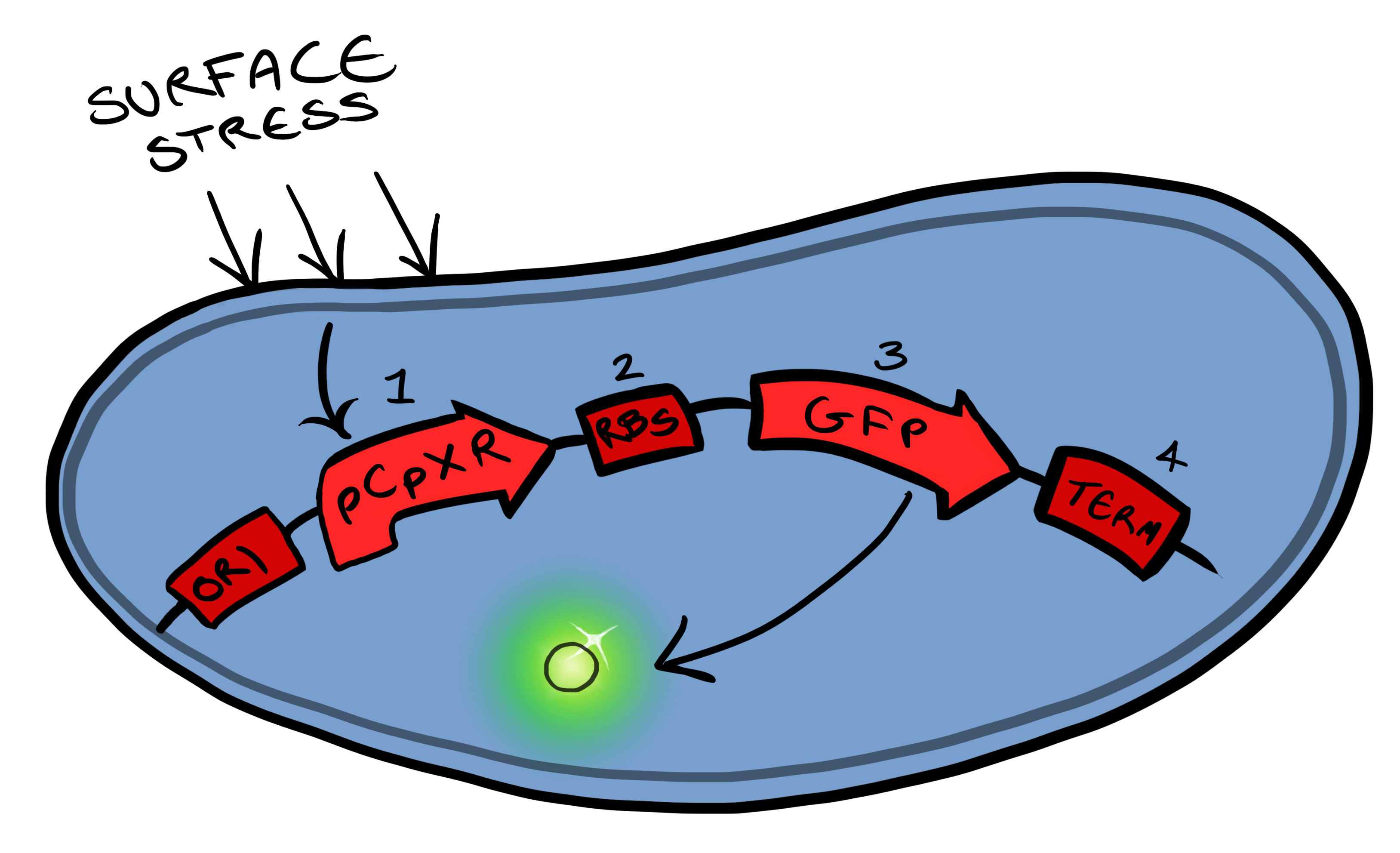
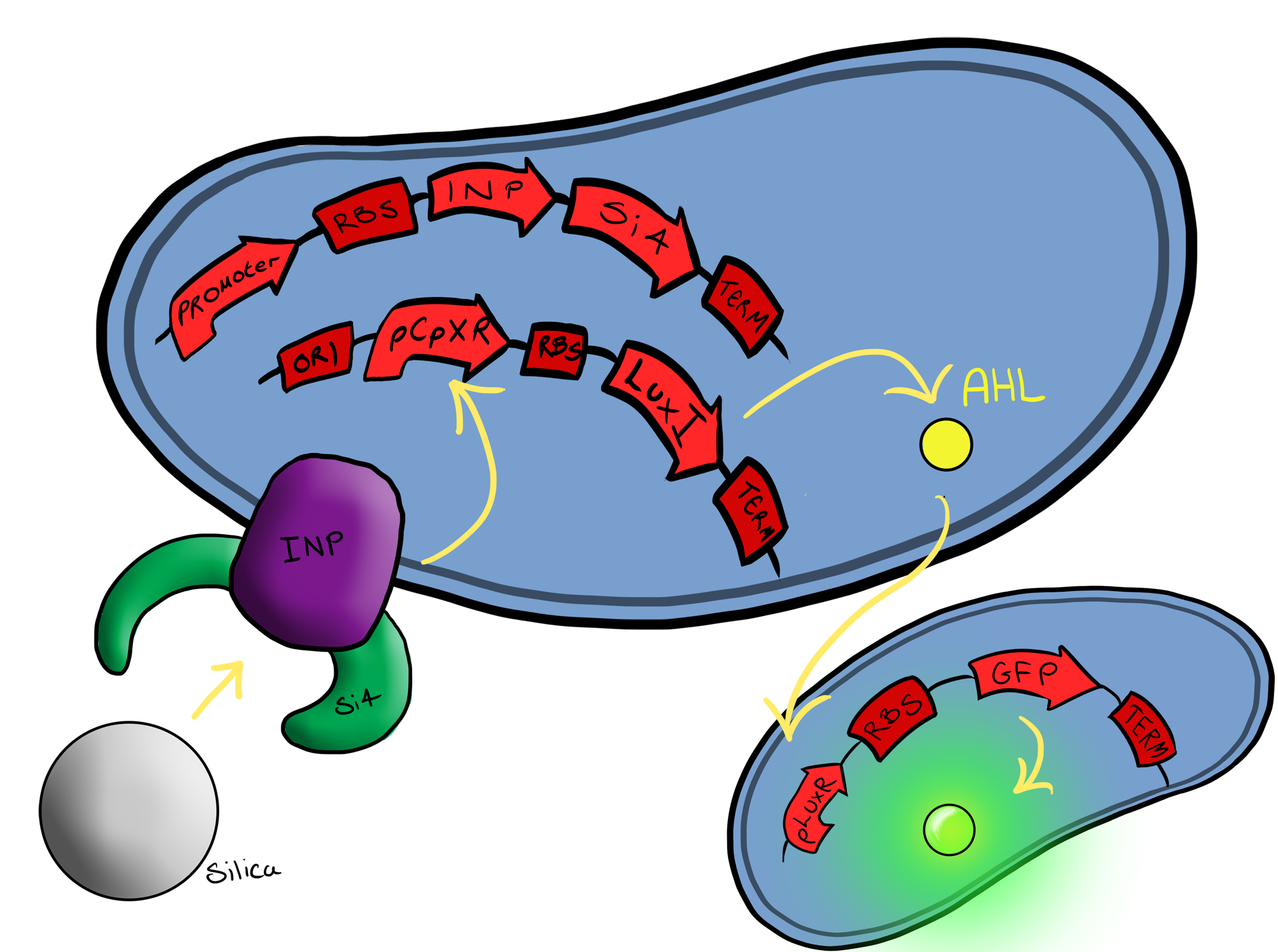
| - | Simultaneously, we will be developing a BioBrick for physical attachment and detection of particles. This will work using Ice Nucleation Protein to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of | + | Simultaneously, we will be developing a BioBrick for physical attachment and detection of particles. This will work using Ice Nucleation Protein to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of pathogen detection. The cells producing this receptor complex will also be constitutively producing a fluorescent reporter of our choice to allow for its detection on silica beads following removal of supernatant and washing. Biobricks we will be using: |
| - | BBa_K081012 Green fluorescent protein generator | + | BBa_K081012 for the Green fluorescent protein generator which contain a ribosome binding site and terminator. |
| - | BBa_K523008 Ice nucleation protein | + | BBa_K523008 is the Ice nucleation protein any sequence added to the C-terminus of the INP will be transported to cell membrane and be displayed on the surface of the cell. |
| - | BBaa_B0015 | + | BBaa_B0015, which is a double terminator, |
| - | BBa_B0034 | + | BBa_B0034 a strong ribosome binding site and finally |
| - | BBa_J23119 Constitutive promoter | + | BBa_J23119 a Constitutive promoter |
| - | Again, fluorescent reporters | + | Again, fluorescent reporters are used for visual purposes in characterisation experiments, but this time the focus of Biosystem 2 is on the adhesion proteins being used. |
<br> | <br> | ||
==Phase II== | ==Phase II== | ||
| Line 84: | Line 84: | ||
===Biosystem 3=== | ===Biosystem 3=== | ||
[[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | [[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | ||
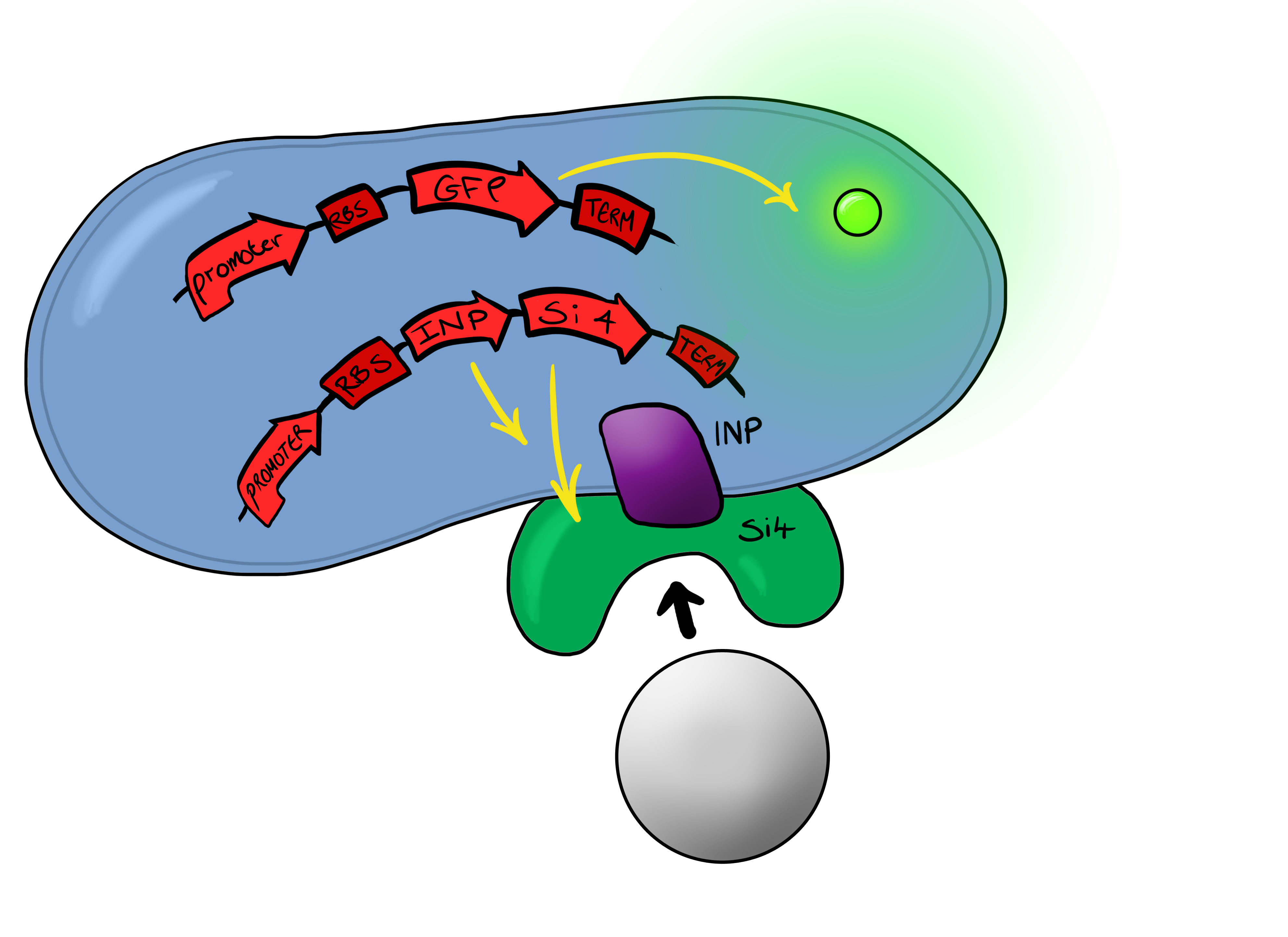
| + | Biosystem 3 brings together BS1 and BS2 in one plasmid. The genes that code for the INP and the si4 binding domain are now located on the same plasmid as the CpxP promoter and the GFP generator. The effect of this is that when the silcon bead binds to the Si4 binding domain connected to the INP protein and causes surface stress the CpxP promoter will be induced. This will cause GFP to be produced and this will act as a visible signal that the 'pathogen' has been detected. | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
==The Cpx Pathway== | ==The Cpx Pathway== | ||
This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | ||
| + | <br style="clear:both"/> | ||
| + | ===A brief explanation of the Cpx Pathway=== | ||
| + | The Cpx pathway responds to various membrane stress; it is a two component signal transduction pathway. The first component is CpxA, an inner membrane protein which, when bound to CpxP (an inhibitor molecule), in inactive. When CpxP is unbound from CpxA, CpxA autophosphorylates itself. A kinase enzyme then phosphorylates the CpxR, the second component in the pathway, which then binds to the Cpx promoter and activates transcription. | ||
| + | There is still a lot of debate regarding this pathway and many different theories but it is thought that membrane stress causes the misfolding of proteins (pili being the main contributor) causes misfolded subunits to accumulate in the periplasm and titrate the CpxP inhibitor molecule off the CpxA and activate the pathway. | ||
}} | }} | ||
Revision as of 13:32, 17 September 2013
 "
"