Team:NYMU-Taipei/Modeling/MainParts
From 2013.igem.org
Mastershot (Talk | contribs) |
Mastershot (Talk | contribs) |
||
| Line 403: | Line 403: | ||
<html> | <html> | ||
<div lang="latex" class="equation"> | <div lang="latex" class="equation"> | ||
| - | \frac{d[ethanol]}{dt}= | + | \frac{d[ethanol]}{dt}=Kpyruvateacetaldehyde\timesKacetaldehydeethanol\times\frac{ [PDC]^{nPDC} }{ KdPDC^{nPDC}+[PDC] }\times\frac{ [ADH]^{nADH} }{ KdADH^{nADH}+[ADH] }\times[pyruvate]-Km\times\frac{ [ADH]^{nADH} }{ KdADH^{nADH}+[ADH]^{nADH} }\times[ethanol] |
</div> | </div> | ||
</html> | </html> | ||
Revision as of 21:28, 27 September 2013


Function of the parts:
- circuit regulators:
- LacIregulatedpromoter(pLac) : when LacI exists, it will bind to LacIregulatedpromoter(pLac) and represses the promoter.
- pLux/cIhybridpromoter(plux/cI) : when luxR/AHL exists, it will open pLux/cIhybridpromoter(plux/cI), while cI will repress the promoter. What’s more, plux/cI is cI-dominant, which means when cI exists, the hybrid promoter will be repressed whether luxR/AHL exists or not.
- Circuit regulation:
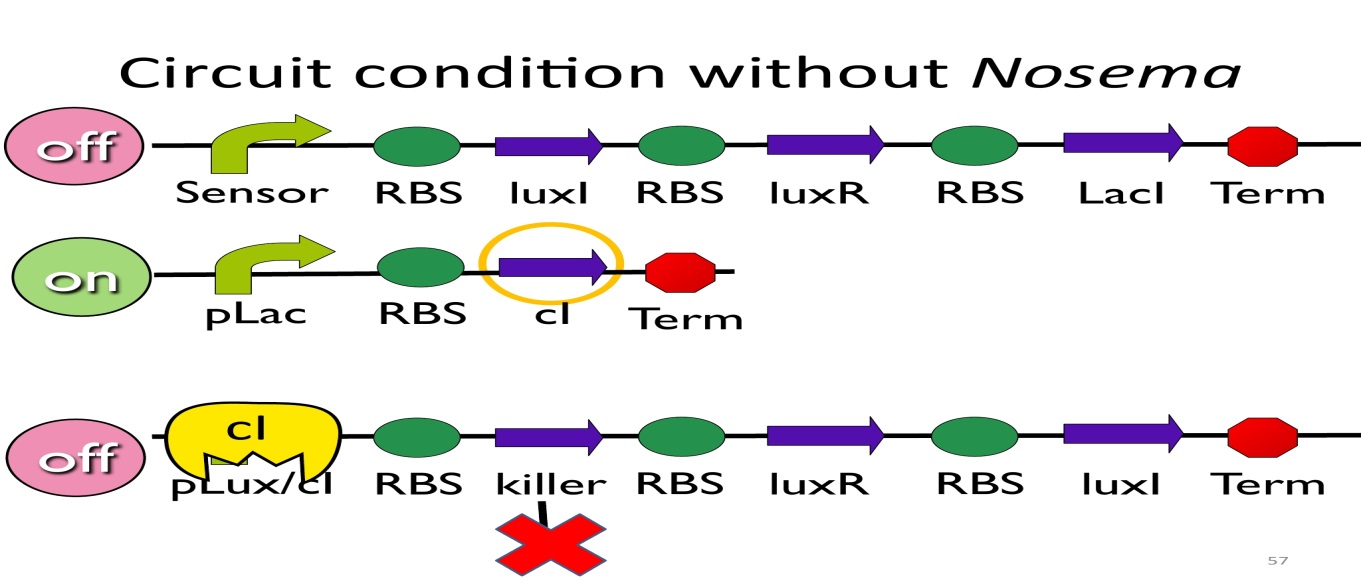
- Circuit condition without Nosema:
Without NosemaCeranae, the first circuit will not open, and thus, no LacI will be produced. Since there is no LacI binding to pLac, pLac will not be repressed, which means cI will be produced. After that, cIwill bind to pLux/cI-hybridpromoter, leading the third circuit to be off.
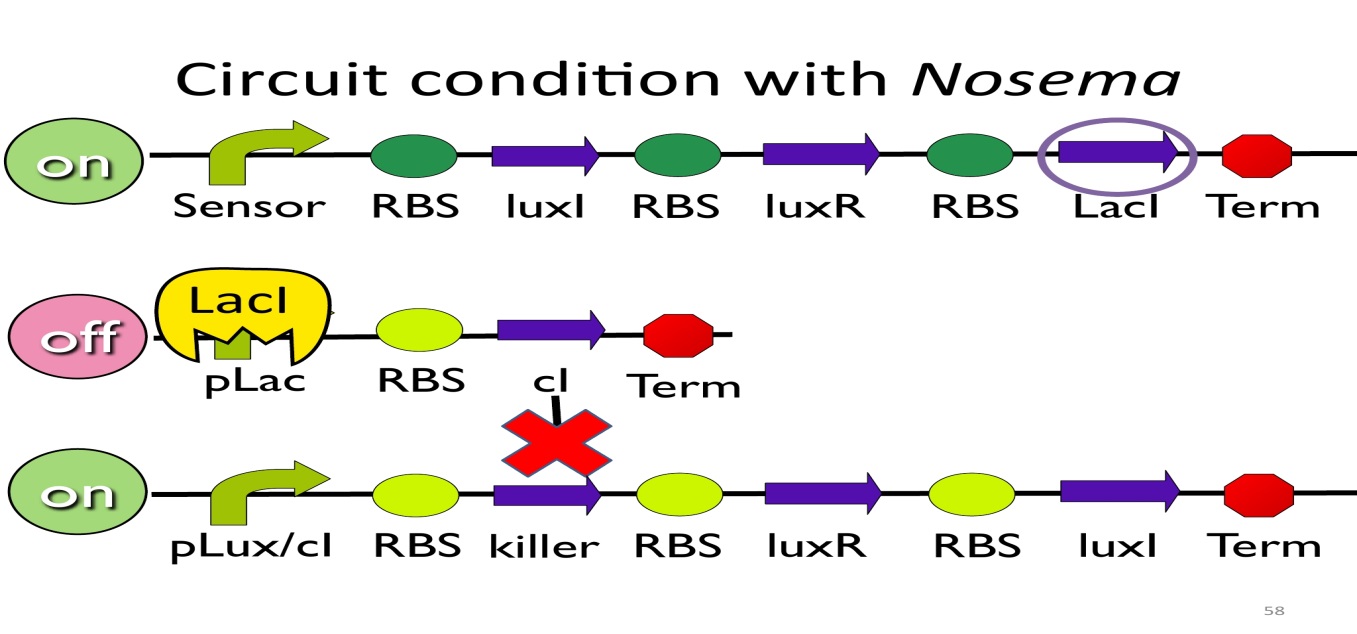
- Circuit condition with Nosema:
When NosemaCeranae infects the bee, bee’s immune system will be activated, leading to the concentration of ROS(reactive oxygen species) to be high. In response, E. Kobee will enhance the production of oxyR, which binds to transcription binding site ahead of trxC (an oxyR-activated promoter; namely, the sensor), leading to the first circuit to be on.
Since the first circuit is on, the downstream LacI gene will be produced and bind to pLac, leading to the second circuit to be off.
After the second circuit is closed, no cI is produced, leading to the third circuit to be on (no repressor at all). And then, killer protein will be produced and kill NosemaCeranae.
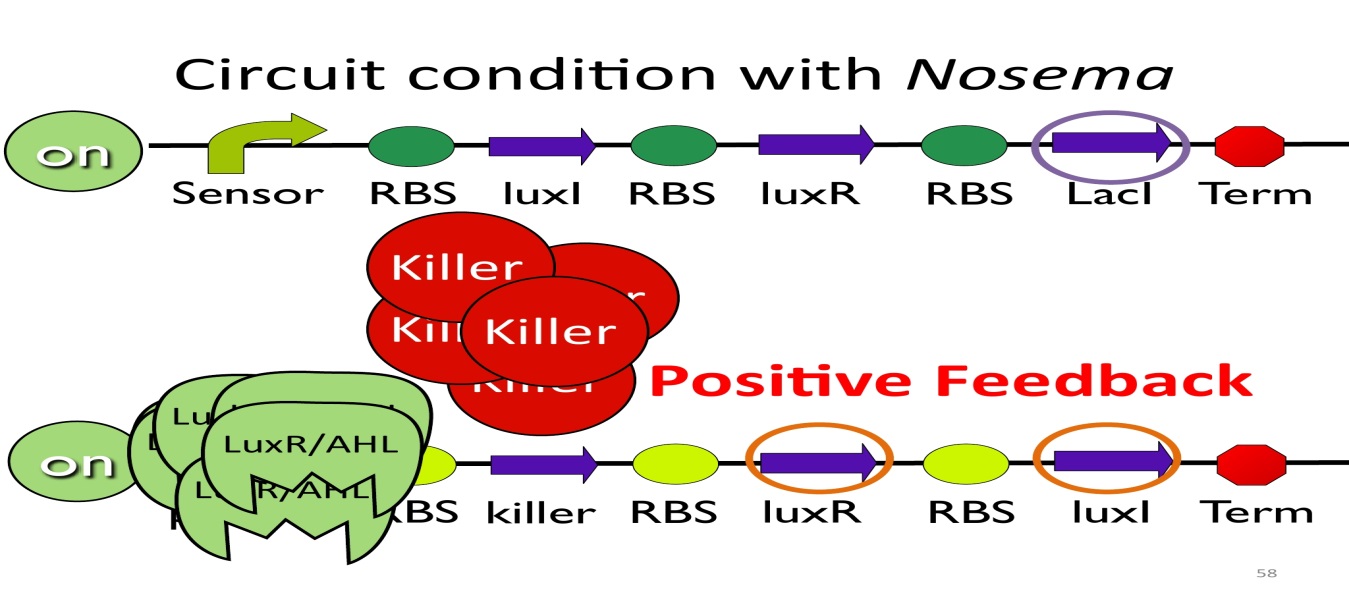
- Positive feedback:
Besides the LuxI and LuxR(which then form a complex called luxR/AHL) produced by first circuit, the third circuit will also produce luxR/AHL, which acts as a positive feedback and strongly enhances the production of killer protein.
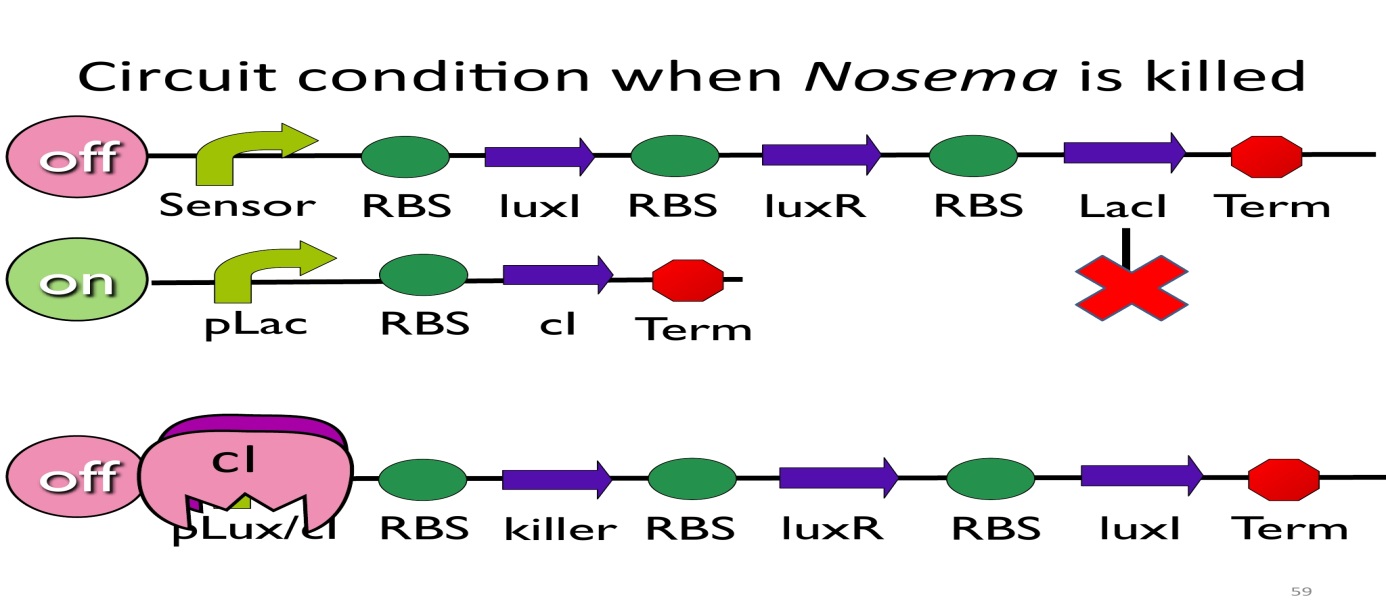
- CircuitconditionwhenNosema is killed:
After Nosema is killed, the sensor will be off and no lacI will be produced. Without LacI, the pLac will not be repressed, and the second circuit will be on, leading to the production of cI. After cI binds topLux/cIhybridpromoter, the third circuit to be off.
The overall circuit will switch to the beginning stage when there is no Nosema.
The purpose of this modeling is:
- To know how much time it needs from sensing to killing the Nosema after infection
- To know whether the pathway is effectiveor not
- To know the range of killer protein concentration(from the minimal concentration which Is effective to kill Nosema to the maximal concentration which will not do harm to the bees)
It is assumed that AHL is abundant and thus the formation of AHL2LuxR is determined only by the concentration of LuxR; CI’s mechanism of binding to the promoter is assumed to act as hill effect form; kill protein will sustain a period of time before it is degraded naturally without any other types of degration.
Equation1:
KdLacI = dissociation constant of CI
nLacI = Hill coefficient of LacI
PoPSpLac = promoter strength of pLac
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNACI production rate and when it can reach the level to translate the desired concentration.
Equation2:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of lacI.
Equation3:
PoPSconstitutive= promoter strength of constitutive promoter (J23102)
N = number of plasmid in a single cell
V = volume of a cell
kdegmRNA = degrading constant of sensor promoter mRNA
The aim of the equation is to know the production rate ofmRNAoxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.
Equation4:

KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant ofcI
ncI = Hill coefficient ofCi
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of LuxI.
Equation5:

KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant ofcI
ncI = Hill coefficient ofCi
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of LuxR.
Equation6:

KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant ofcI
ncI = Hill coefficient ofCi
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of kill protein production rate and when it can reach the level ofthe desired concentration.
Equation7:

KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of T7 polymerase production rate and when it can reach the level to translate enough T7 polymerase.
Equation8:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC.
Equation9:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme ADH production rate and when it can reach the level to translate enough ADH.
Equation10:
RBS = binding site strength
KdegCI = degrading constant of CI
The aim of the equation is to know the production rate of CI and when it can reach the concentration to repress LuxCI hybrid promoter.
Equation11:
RBS = binding site strength
KdegLacI = degrading constant of LacI
The aim of the equation is to knowLacI production rate and when it can reach the concentration to repress promoter LacI
Equation12:
RBS = binding site strength
KAHL = LuxIAHL rate constant
KdegLuxI = degrading constant of LuxI
The aim of the equation is to know the production rate of LuxI concerning the LuxIAHL reaction and LuxI degrading.
Equation13:
RBS = binding site strength
KmAHL =AHLLuxI rate constant
KdegLuxR= degrading constant of LuxR
KonAHL = 2AHL + LuxR(AHL)2/LuxR complex rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of LuxR concerning the 2AHL + LuxR↔(AHL)2/LuxR reaction , AHL→LuxI reaction, and LuxR, AHL degrading.
Equation14:
KAHL = LuxIAHL rate constant
KonAHL = 2AHL + LuxR(AHL)2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex2AHL + LuxR rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of AHL concerning the LuxI→AHL reaction, 2AHL + LuxR↔(AHL)2/LuxRreaction and AHL degrading.
Equation15:
RBS = binding site strength
KdegT7 = degrading constant of T7
The aim of the equation is to know the threshold concentration of oxyR to conquer the terminal and when T7 polymerase can reach the required concentration to activate T7 promoter.
Equation16:
RBS = binding site strength
KdegPDC = degrading constant of PDC
The aim of the equation is to know PDC production rate and when it can reach the concentration of ethanol pathway equilibrium.
Equation17:
RBS = binding site strength
KdegADH = degrading constant of ADH
The aim of the equation is to know ADH production rate and when it can reach the concentration of ethanol pathway equilibrium.
Equation18:
KonAHL = 2AHL + LuxR(AHL)2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex2AHL + LuxR rate constant
KonAHL = 2AHL + LuxR(AHL)2/LuxR complex rate constant KoffAHL = (AHL)2/LuxR complex2AHL + LuxR rate constant
Equation19:
RBS = binding site strength
Kdegkill = degrading constant of kill
The aim of the equation is to knowkiller protein production rate and when it can reach the effective concentration to killNosema.
Equation20:
Kpyruvateacetaldehyde= pyruvate→acetaldehyde reaction rate constant
Kacetaldehydeethanol= acetaldehyde→ethanol reaction rate constant
Km = ethanol→acetaldehyde reaction rate constant
KdPDC = dissociation constant of PDC
KdADH = dissociation constant of ADH
The aim of the equation is to know ethanol production rate and when it can reach the concentration to kill the spores of NosemaCeranae.
 "
"