Team:Paris Saclay/Notebook/August/27
From 2013.igem.org
(→1 - Electrophoresis of RBS-BphR2 Part I, BphR2 Part II, FNR Part I , FNR Part II, RBS-FNR Part I and PSB1C3) |
(→1 - Electrophoresis of RBS-BphR2 Part I, BphR2 Part II, FNR Part I , FNR Part II, RBS-FNR Part I and pSB1C3) |
||
| Line 91: | Line 91: | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | * pSB1C3 : 2070 bp |
* RBS-BphR2 Part I : 197 bp | * RBS-BphR2 Part I : 197 bp | ||
* BphR2 Part II : 790 bp | * BphR2 Part II : 790 bp | ||
| Line 100: | Line 100: | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We obtain fragment at the right size for RBS-BphR2 Part I, BphR2 Part II, RBS-FNR Part I, FNR Part I, FNR Part II but not for | + | We obtain fragment at the right size for RBS-BphR2 Part I, BphR2 Part II, RBS-FNR Part I, FNR Part I, FNR Part II but not for pSB1C3. We will do a digestion of PSB1C3 by DnpI to clean it. |
|} | |} | ||
Revision as of 15:58, 4 October 2013
Notebook : August 27
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000 and BBa_K1155004
1 - PCR Colony of ligation Pfnr or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3
XiaoJing
|
Transformation of 08/26/13 works. We will do a PCR Colony. |
We took a single colony and resuspend in 10µL H2O.For each Biobrick we did 6 PCR Colony.
Used quantities :
- DNA : 2µL
- Mix : (it was divided in 6 tubes for each promotor with 23µL of mix in each tube)
- Oligo 44 : 17.5µL
- Oligo 43 : 17.5µL
- Buffer Dream Taq : 87.5µL
- dNTP : 17.5µL
- Dream Taq : 7µL
- H2O : 591µL
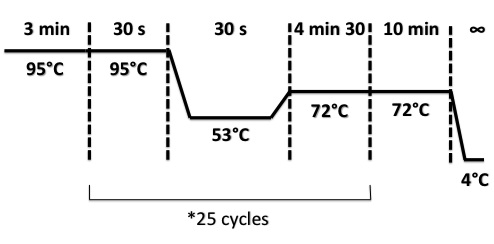
PCR Program :
2 - Electrophoresis of the colony PCR products : Pfnr or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in PSB1C3
XiaoJing
| [[]] |
|
Expected size :
- NirB with RBS-LacZ-Term in pSB1C3 : 3474bp
- NirB with RBS-AmilCP-Term in pSB1C3 : 1029bp
- Pfnr with RBS-LacZ-Term in pSB1C3 : 3380bp
- Pfnr with RBS-AmilCP-Term in pSB1C3 : 935bp
|
We obtain fragments at the right size for NirB with RBS-Amil CP-Term in PSB1C3 in well 12, Pfnr with RBS-LacZ-Term in pSB1C3 in well 14, 15, 18 and 19 and Pfnr with RBS-Amil CP-Term in pSB1C3 in well 20, 22 and 25. Nevertheless, electrophoresis shows that these colonies weren't pure. We will purify them by streaking. |
3 - Streak colonies of NirB with RBS-Amil CP-Term in pSB1C3, Pfnr with RBS-LacZ-Term in pSB1C3 and Pfnr with RBS-Amil CP-Term in pSB1C3 to purify them
XiaoJing
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR, BphR2 proteins
1 - Electrophoresis of RBS-BphR2 Part I, BphR2 Part II, FNR Part I , FNR Part II, RBS-FNR Part I and pSB1C3
XiaoJing
|
Gibson tranformation of the 08/27/13 didn't work. So we did an electrophoresis to check sizes and concentrations of Gibson parts. |
| [[]] |
|
Expected sizes :
- pSB1C3 : 2070 bp
- RBS-BphR2 Part I : 197 bp
- BphR2 Part II : 790 bp
- RBS-FNR Part I : 615 bp
- FNR Part I : 597 bp
- FNR Part II : 200 bp
|
We obtain fragment at the right size for RBS-BphR2 Part I, BphR2 Part II, RBS-FNR Part I, FNR Part I, FNR Part II but not for pSB1C3. We will do a digestion of PSB1C3 by DnpI to clean it. |
2 - Digestion of pSB1C3 by DnpI
XiaoJing
Used quantities :
- pSB1C3 : 17µL
- Buffer : 2µL
- DnpI : 1µL
We keep the digestion for 1h30 at 37°C.
3 - Electrophoresis of the digestion of pSB1C3 by DnpI
XiaoJing
| [[]] |
|
Expected sizes :
- pSB1C3 : 2070 bp
|
We obtain fragment at the right size for pSB1C3. We will purify it. |
| Previous day | Back to calendar | Next day |
 "
"