Team:Freiburg/Notebook/modeling
From 2013.igem.org
| Line 60: | Line 60: | ||
</p> | </p> | ||
<div id="text1"> | <div id="text1"> | ||
| - | <p> | + | <p>65,000 cells per well were seeded. 24 h before transfection.<br> |
The cells had been transfected at 21:30 h. </p> | The cells had been transfected at 21:30 h. </p> | ||
</div> | </div> | ||
| Line 71: | Line 71: | ||
<tr> | <tr> | ||
<td> <b>Figure 1: <b> Transfection Scheme</b> </b><br> | <td> <b>Figure 1: <b> Transfection Scheme</b> </b><br> | ||
| - | Cells were transfected with dCas9-VP16 (transfection one) and as a control with reporter plasmid and junk DNA (transfection two). For repression dCas9-KRAB, trans-activator and SEAP reporter plasmid were transfected (transfection 3). As control reporter and | + | Cells were transfected with dCas9-VP16 (transfection one) and as a control with reporter plasmid and junk DNA (transfection two). For repression dCas9-KRAB, trans-activator and SEAP reporter plasmid were transfected (transfection 3). As control reporter and transactivator only was transfected (transfection 4). |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 92: | Line 92: | ||
<p> | <p> | ||
| - | <br>Every timepoint the | + | <br>Every timepoint the supernatant was taken for SEAP measurement and the cells were lysated with RIPA buffer for quantitative western blotting.<br> |
The standard western blot procedure was done with 20 µl of the cell lysates.<br> | The standard western blot procedure was done with 20 µl of the cell lysates.<br> | ||
| Line 98: | Line 98: | ||
For quantification of the western blot data, a dot blot was done. A serial dilution of anti-HA (200ng/µl)was prepared. <br><br> | For quantification of the western blot data, a dot blot was done. A serial dilution of anti-HA (200ng/µl)was prepared. <br><br> | ||
1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200, 1:6400<br><br> | 1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200, 1:6400<br><br> | ||
| - | After PVDF-membrane activation in methanol and a short incubation in tranfer buffer, 2 µl of each dilution have been | + | After PVDF-membrane activation in methanol and a short incubation in tranfer buffer, 2 µl of each dilution have been dropped onto the membrane. After that the membrane was incubated at room temerature (1h) for drying.<br> |
The detection with the secondary anti-body was done similar to the treatment of the dCas9 blots.<br><br> | The detection with the secondary anti-body was done similar to the treatment of the dCas9 blots.<br><br> | ||
</p> | </p> | ||
| Line 118: | Line 118: | ||
<tr> | <tr> | ||
<td> <b>Figure 2: <b> Western Blot</b> </b><br> | <td> <b>Figure 2: <b> Western Blot</b> </b><br> | ||
| - | The dCas9 fusion protein possesses a HA-tag and could so be | + | The dCas9 fusion protein possesses a HA-tag and could so be detected with an anti-HA antibody. The first two blots show dCas9-KRAB. The particular timepoints are labelled above the band. The same pertains for the two dCas9-VP16 blots. As negative control (c), the lysate of timepoint 6 of transfection 3 and 4 has been used. |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 132: | Line 132: | ||
<tr> | <tr> | ||
<td> <b>Figure 3: <b> Dot Blot </b> </b><br> | <td> <b>Figure 3: <b> Dot Blot </b> </b><br> | ||
| - | The dots are labelled with their | + | The dots are labelled with their dilution rate. Only the labelled dots were used for quanitfication. The blots of the first line have been illuminated under the same conditions as the dCas9-KRAB blots, the one of the second line under conditions as the dCas9-VP16 blots. |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 138: | Line 138: | ||
</div> | </div> | ||
| - | <p>For the quatification, the dot intensity has been analysed with Image J. The data was used to generate a | + | <p>For the quatification, the dot intensity has been analysed with Image J. The data was used to generate a calibration curve. The intesities of the dCas9 bands were compared with the calibration curve, so that the amount of dCas9 per band could be defined.</p> |
</body> | </body> | ||
</html> | </html> | ||
Revision as of 03:09, 5 October 2013

Modeling Notebook
65,000 cells per well were seeded. 24 h before transfection.
The cells had been transfected at 21:30 h.
 |
| Figure 1: Transfection Scheme Cells were transfected with dCas9-VP16 (transfection one) and as a control with reporter plasmid and junk DNA (transfection two). For repression dCas9-KRAB, trans-activator and SEAP reporter plasmid were transfected (transfection 3). As control reporter and transactivator only was transfected (transfection 4). |
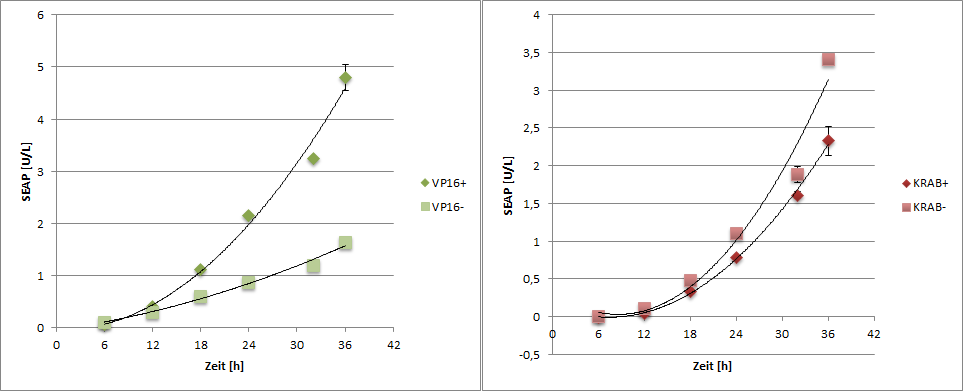
To generate time dependent data, one well was treated every 6 h.
t0= 01:30
t1= 07:30
t2= 13:30
t3= 19:30
t4= 01:30
t5= 07:50
t6= 13:30
Every timepoint the supernatant was taken for SEAP measurement and the cells were lysated with RIPA buffer for quantitative western blotting.
The standard western blot procedure was done with 20 µl of the cell lysates.
For quantification of the western blot data, a dot blot was done. A serial dilution of anti-HA (200ng/µl)was prepared.
1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200, 1:6400
After PVDF-membrane activation in methanol and a short incubation in tranfer buffer, 2 µl of each dilution have been dropped onto the membrane. After that the membrane was incubated at room temerature (1h) for drying.
The detection with the secondary anti-body was done similar to the treatment of the dCas9 blots.
Results
 |
| Figure 2: Western Blot The dCas9 fusion protein possesses a HA-tag and could so be detected with an anti-HA antibody. The first two blots show dCas9-KRAB. The particular timepoints are labelled above the band. The same pertains for the two dCas9-VP16 blots. As negative control (c), the lysate of timepoint 6 of transfection 3 and 4 has been used. |
 |
| Figure 3: Dot Blot The dots are labelled with their dilution rate. Only the labelled dots were used for quanitfication. The blots of the first line have been illuminated under the same conditions as the dCas9-KRAB blots, the one of the second line under conditions as the dCas9-VP16 blots. |
For the quatification, the dot intensity has been analysed with Image J. The data was used to generate a calibration curve. The intesities of the dCas9 bands were compared with the calibration curve, so that the amount of dCas9 per band could be defined.
 "
"




