Team:NYMU-Taipei/Project/Inhibition/Prohibition
From 2013.igem.org
| Line 16: | Line 16: | ||
==Experiment== | ==Experiment== | ||
| - | To check whether FimH really works in our ''Bee. coli'', we fused RFP and FimH protein([[http://parts.igem.org/Part:BBa_K1104101|BBa_K1104101]]), so that when FimH binds to the mannose polymer, we can easily observe through seeing the red fluorescence.<br> | + | To check whether FimH really works in our ''Bee. coli'', we fused RFP and FimH protein mannose-binding domain([[http://parts.igem.org/Part:BBa_K1104101|BBa_K1104101]]), so that when FimH binds to the mannose polymer, we can easily observe through seeing the red fluorescence.We will call this fusion protein RFP-FimH on the following text.<br> |
| + | [[File:NYMU RFP-FimH(XP).png|right|thumb|100px|ab]] | ||
===Circuit Design=== | ===Circuit Design=== | ||
| - | |||
Through blind ligation, we insert the 22nd to 179th amino acid of FimH protein gene in front the stop codon of the RFP coding sequence, in order to form a protein fusion. After cloning the FimH gene and backbone, we have done blind ligation to fuse RFP and FimH protein into one. The picture on the right is the circuit design for our experiment.<br> | Through blind ligation, we insert the 22nd to 179th amino acid of FimH protein gene in front the stop codon of the RFP coding sequence, in order to form a protein fusion. After cloning the FimH gene and backbone, we have done blind ligation to fuse RFP and FimH protein into one. The picture on the right is the circuit design for our experiment.<br> | ||
[[File:NYMU_RFP-FimH cell.jpg|left|thumb|200px|ab]] | [[File:NYMU_RFP-FimH cell.jpg|left|thumb|200px|ab]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
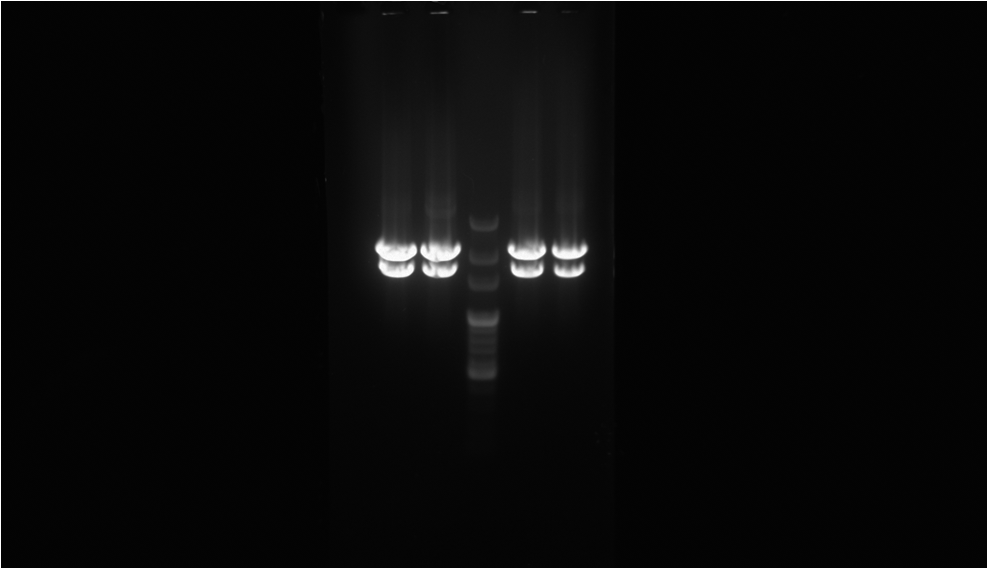
To proof that the protein fusion actually express in the ''E. coli'' we have done digestion on XbaI and PstI cutting site to see if the length of our biobrick is right. And through observing the fluorescence emitted by the ''E. coli'' we can be sure that we have produced the protein that we want.The result is as the picture on the left.<br> | To proof that the protein fusion actually express in the ''E. coli'' we have done digestion on XbaI and PstI cutting site to see if the length of our biobrick is right. And through observing the fluorescence emitted by the ''E. coli'' we can be sure that we have produced the protein that we want.The result is as the picture on the left.<br> | ||
| - | === | + | |
| + | |||
| + | |||
| + | |||
| + | ===RFP-FimH functional test=== | ||
After cloning mngB gene into ''E. coli'', it is a must to examine whether our part is properly combined and well-functioned. Therefore we decide to extract alpha-mannosidase from the cytoplasm of the ''E. coli'' and test its function by directly interact with 2-O-(6-phospho-α-D-mannosyl)-D-glycerate. With the aid of HPLC, we can see if it can successfully turn it into α-D-mannose-6-phosphate and D-glycerate. | After cloning mngB gene into ''E. coli'', it is a must to examine whether our part is properly combined and well-functioned. Therefore we decide to extract alpha-mannosidase from the cytoplasm of the ''E. coli'' and test its function by directly interact with 2-O-(6-phospho-α-D-mannosyl)-D-glycerate. With the aid of HPLC, we can see if it can successfully turn it into α-D-mannose-6-phosphate and D-glycerate. | ||
====Experimental process is listed below==== | ====Experimental process is listed below==== | ||
Revision as of 18:12, 27 October 2013


Contents |
Prohibiting Sprouting
Introduction
In order to deter the invasion of Nosema ceranae, the first step we take is to fully understand its life cycle which completes by bees. The picture beside indicates the relation betweenNosema ceranae stage and the place it grows. By observing these information, we can obviously analyze the advantages and drawbacks at each stage, as well as decide which stage is the best and most attainable for us to weaken the transmission ability of Nosema ceranae.
Background
Stop it at the very begining!!!
After N. ceranae invades epithelial cells of a bee's midgut, it will soon reproduce in two days and new generation appears. Since its lifecycle is so short that there is no time for biological methods to react and inhibit the development of the pathogen, we choose to inhibit it before entering bees' epithelial cells, which will be before N. ceranae’s germination. When N. ceranae's spore moves to bees' midgut, it will start to germinate and develop a structure called polar filament. This structure will then bind to epithelial cells of bees' midgut and poke a hole at the membrane of an epithelial cell, causing the bee eventually dead. In order to inhibit polar filament's development, we find out that there is a protein called PTP1, a protein on the surface of the polar filament, is highly relevant to the binding of the polar filament and epithelial cells. It is also highly believed that obstructing the production of PTP1 can stop the invasion of N. ceranae into the epithelial cell of the bee's midgut. So we decide to suppress the growth of the polar filament by obstruct the production of PTP1. In the research that is done by other scientist, they have found out that in the process of PTP1 formation a mannose polymer will bind to the protein. We initially decided to use mannosidase as our weapon to stop the invasion, however, we have considered the efficiency of how fast the enzyme can degrade the polymer, also due to the difficulty to test its function, we changed our weapon into FimH, which is a mannose binding protein. In this way, at the appearance of the polar filament our protein can immediately block PTP1 and stop the invasion of N. ceranae.
FimH
FimH protein, a protein expressed in Escherichia coli K-12 substr MG1655, has a mannose-binding site from the 22nd to 179th amino acid. FimH protein is originally located on the tip of the fimbria of the E. coli, which will adhere to the mannose on the host's cell membrane, allowing the bacteria to colonize various host tissues. In our project we managed to use it to bind the mannose polymer, which serves as a sticky end to bind the host cell, on top of the PTP1, in order to stop the adhesion between the polar filament and the epithelial cell of the bee's midgut.
Circuit Design
Experiment
To check whether FimH really works in our Bee. coli, we fused RFP and FimH protein mannose-binding domain(BBa_K1104101), so that when FimH binds to the mannose polymer, we can easily observe through seeing the red fluorescence.We will call this fusion protein RFP-FimH on the following text.
Circuit Design
Through blind ligation, we insert the 22nd to 179th amino acid of FimH protein gene in front the stop codon of the RFP coding sequence, in order to form a protein fusion. After cloning the FimH gene and backbone, we have done blind ligation to fuse RFP and FimH protein into one. The picture on the right is the circuit design for our experiment.
To proof that the protein fusion actually express in the E. coli we have done digestion on XbaI and PstI cutting site to see if the length of our biobrick is right. And through observing the fluorescence emitted by the E. coli we can be sure that we have produced the protein that we want.The result is as the picture on the left.
RFP-FimH functional test
After cloning mngB gene into E. coli, it is a must to examine whether our part is properly combined and well-functioned. Therefore we decide to extract alpha-mannosidase from the cytoplasm of the E. coli and test its function by directly interact with 2-O-(6-phospho-α-D-mannosyl)-D-glycerate. With the aid of HPLC, we can see if it can successfully turn it into α-D-mannose-6-phosphate and D-glycerate.
Experimental process is listed below
1.Use Continuous High Pressure Cell Disrupter to crush E. coli that can produce alpha-mannosidase
2.Filter the product and save the filtrate in an eppendorf
3.Add 2-O-(6-phospho-α-D-mannosyl)-D-glycerate into eppendorf
4.Incubate it at 25℃ for 30 minutes.
5.Run HPLC and compare it with the two standard line of only 2-O-(6-phospho-α-D-mannosyl)-D-glycerate, and with only the extraction liquid of E. coli cytoplasm
6.Compare the area of the data lines with the area of the standard line to calculate the efficiency of the alpha-mannosidase produced by E. coli
Reference
1. EcoCyc: Encyclopedia of Escherichia coli K-12 Genes and Metabolism [http://ecocyc.org/ECOLI/NEW-IMAGE?type=ENZYME&object=EG13236-MONOMER EcoCyc/ alpha mannosidase]
2. XU, Y., & WEISS, L. (August 01, 2005). The microsporidian polar tube: A highly specialised invasion organelle. International Journal for Parasitology, 35, 9, 941-953.
3. Xu, Yanji, Takvorian, Peter M., Cali, Ann, Orr, George, & Weiss, Louis M. (n.d.). Glycosylation of the Major Polar Tube Protein of Encephalitozoon hellem, a Microsporidian Parasite That Infects Humans. American Society for Microbiology.
4. Xu, Yanji, Takvorian, Peter M., Cali, Ann, Orr, George, & Weiss, Louis M. (n.d.). Glycosylation of the Major Polar Tube Protein of Encephalitozoon hellem, a Microsporidian Parasite That Infects Humans. American Society for Microbiology.
 "
"