Team:Paris Saclay/Notebook/August/27
From 2013.igem.org
(Difference between revisions)
(→summary) |
(→summary) |
||
| Line 6: | Line 6: | ||
*We got colonies after the transformation of E.coli with the ligation mix "PnirB_PSB1C3 and RBS_LacZ_Term" | *We got colonies after the transformation of E.coli with the ligation mix "PnirB_PSB1C3 and RBS_LacZ_Term" | ||
promoter fnr(activator)nirB plus RBS_AmilCP+Term_PSB1C3 | promoter fnr(activator)nirB plus RBS_AmilCP+Term_PSB1C3 | ||
| - | + | "Pndh*_PSB1C3 plus RBS_LacZ+Term" | |
| - | + | "Pndh*_PSB1C3 plus RBS_AmilCP+Term" | |
| - | so we | + | so we tested for correct ligation with PCR on colonies |
| - | * We | + | * We did not get any colonies for the Gibson assembly so we analyzed by gel electrophoresis the parts used for the Gibson assembly to verify their sizes and concentrations. |
| - | * Digestion | + | * Digestion DnpI purification of the the PSB1C3 clean for Gibson and electrophoresis of it . |
<br> | <br> | ||
Revision as of 12:10, 21 September 2013
Contents |
Notebook : August 27
summary
- We got colonies after the transformation of E.coli with the ligation mix "PnirB_PSB1C3 and RBS_LacZ_Term"
promoter fnr(activator)nirB plus RBS_AmilCP+Term_PSB1C3 "Pndh*_PSB1C3 plus RBS_LacZ+Term" "Pndh*_PSB1C3 plus RBS_AmilCP+Term" so we tested for correct ligation with PCR on colonies
- We did not get any colonies for the Gibson assembly so we analyzed by gel electrophoresis the parts used for the Gibson assembly to verify their sizes and concentrations.
- Digestion DnpI purification of the the PSB1C3 clean for Gibson and electrophoresis of it .
lab work
- A.aero/anaerobic regulation system
- 1.Colony PCR on e.coli with FNR Promoter plus RBS_LacZ+Term_PSB1C3 or RBS_AmilCP+Term_PSB1C3 for 8 colonies of each
Xiaojing
- A1-8: promoter fnr(activator)nirB plus RBS_LacZ+Term_PSB1C3
- B1-8: promoter fnr(activator)nirB plus RBS_AmilCP+Term_PSB1C3
- C1-8: promoter fnr(repressor) plus RBS_LacZ+Term_PSB1C3
- D1-8: promoter fnr(repressor) plus RBS_AmilCP+Term_PSB1C3
- Picking of 32 colonies
- Preparation Master mix
- H2O : 490µL
- dNTP : 17.5µL
- VR primer : 17.5µL
- VF2 primer : 17.5µL
- DreamTaq buffer 10x : 87.5µL
- DreamTaq enzyme : 7µL
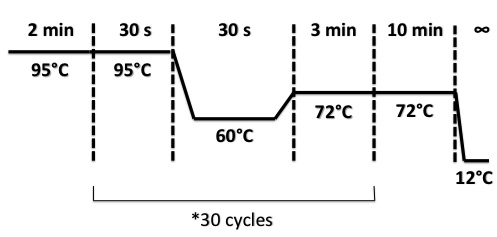
Protocol : Colony PCR PCR Program :
2 - Gel electrophoresis of the colony PCR products
| [[]] |
|
Expected size : 3583bp
A - Aerobic/Anaerobic regulation system / B - PCB sensing system
1 -Gel electrophoresis of parts of Gibson assembly.
| [[]] |
|
Now we do a Digestion Dnp1 purification of the the PSB1C3 clean for Gibson.
| PSB1C3 clean for Gibson | 17µl |
| enzyme Dnp1 | 1µl |
| buffer | 2µl |
| total | 20µl |
- Incubate at 37°C for 1h30mins.
2 - Gel purification of the PSB1C3 Clean for Gibson
2.1 - Migration of 20µl PSB1C3 Clean for Gibson digested by Dnp1 '
| [[]] |
|
| Previous week | Back to calendar | Next day |
 "
"