Team:Paris Saclay/Notebook/August/27
From 2013.igem.org
(Difference between revisions)
(→summary) |
|||
| Line 119: | Line 119: | ||
|[[Team:Paris Saclay/Notebook/July/2|<big>Next day</big>]] | |[[Team:Paris Saclay/Notebook/July/2|<big>Next day</big>]] | ||
|} | |} | ||
| + | |||
| + | |||
| + | {{Team:Paris_Saclay/incl_fin}} | ||
| + | |||
| + | {{Team:Paris_Saclay/incl_debut_generique}} | ||
| + | |||
| + | ='''Notebook : August 5'''= | ||
| + | |||
| + | =='''Lab work'''== | ||
| + | |||
| + | ==='''A - Aerobic/Anaerobic regulation system'''=== | ||
| + | |||
| + | ===='''Objective : characterize Bba_K1155004, Bba_K1155005, Bba_K1155006'''==== | ||
| + | |||
| + | ===='''1 - Colony PCR of ligation Pfnr or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in PSB1C3'''==== | ||
| + | |||
| + | XiaoJing | ||
| + | |||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DE;" | | ||
| + | Transformation of 08/26/13 works. We will do a PCR Colony. | ||
| + | |} | ||
| + | |||
| + | COLONIES PIQUEES DANS 10µL d'eau par Tube !!!!!!!!!!!!! | ||
| + | |||
| + | Used quantities : | ||
| + | * DNA : 2µL | ||
| + | * Mix : (it was divided in 6 tubes for each promotor with 23µL of mix in each tube) | ||
| + | ** Oligo 44 : 17.5µL | ||
| + | ** Oligo 43 : 17.5µL | ||
| + | ** Buffer Dream Taq : 87.5µL | ||
| + | ** dNTP : 17.5µL | ||
| + | ** Dream Taq : 7µL | ||
| + | ** H2O : 591µL | ||
| + | |||
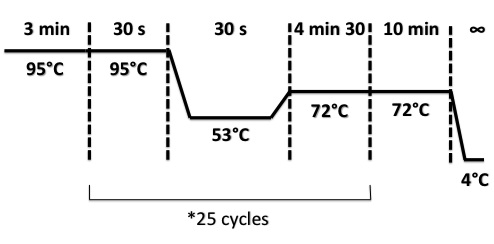
| + | PCR Program : | ||
| + | |||
| + | [[File:PsPCR2708.jpg|400px]] | ||
| + | |||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Revision as of 18:00, 21 September 2013
Contents |
Notebook : August 27
summary
- We got colonies after the transformation of E.coli with the ligation mix "PnirB_PSB1C3 and RBS_LacZ_Term"
"PnirB_PSB1C3 plus RBS_AmilCP+Term" "Pndh*_PSB1C3 plus RBS_LacZ+Term" "Pndh*_PSB1C3 plus RBS_AmilCP+Term" so we tested for correct ligation with PCR on colonies
- We did not get any colonies for the Gibson assembly so we analyzed by gel electrophoresis the parts used for the Gibson assembly to verify their sizes and concentrations.
- Digestion DnpI purification of the the PSB1C3 clean for Gibson and electrophoresis of it .
lab work
- A.aero/anaerobic regulation system
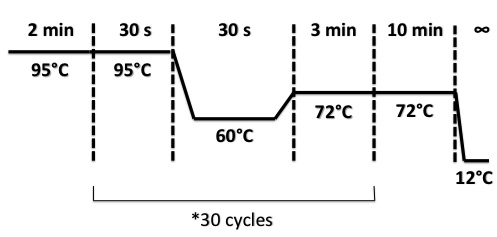
- 1.Colony PCR on e.coli with FNR Promoter plus RBS_LacZ+Term_PSB1C3 or RBS_AmilCP+Term_PSB1C3 for 8 colonies of each
Xiaojing
- A1-8: promoter fnr(activator)nirB plus RBS_LacZ+Term_PSB1C3
- B1-8: promoter fnr(activator)nirB plus RBS_AmilCP+Term_PSB1C3
- C1-8: promoter fnr(repressor) plus RBS_LacZ+Term_PSB1C3
- D1-8: promoter fnr(repressor) plus RBS_AmilCP+Term_PSB1C3
- Picking of 32 colonies
- Preparation Master mix
- H2O : 490µL
- dNTP : 17.5µL
- VR primer : 17.5µL
- VF2 primer : 17.5µL
- DreamTaq buffer 10x : 87.5µL
- DreamTaq enzyme : 7µL
Protocol : Colony PCR PCR Program :
2 - Gel electrophoresis of the colony PCR products
| [[]] |
|
Expected size : 3583bp
A - Aerobic/Anaerobic regulation system / B - PCB sensing system
1 -Gel electrophoresis of parts of Gibson assembly.
| [[]] |
|
Now we do a Digestion Dnp1 purification of the the PSB1C3 clean for Gibson.
| PSB1C3 clean for Gibson | 17µl |
| enzyme Dnp1 | 1µl |
| buffer | 2µl |
| total | 20µl |
- Incubate at 37°C for 1h30mins.
2 - Gel purification of the PSB1C3 Clean for Gibson
2.1 - Migration of 20µl PSB1C3 Clean for Gibson digested by Dnp1 '
| [[]] |
|
| Previous week | Back to calendar | Next day |
Notebook : August 5
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize Bba_K1155004, Bba_K1155005, Bba_K1155006
1 - Colony PCR of ligation Pfnr or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in PSB1C3
XiaoJing
|
Transformation of 08/26/13 works. We will do a PCR Colony. |
COLONIES PIQUEES DANS 10µL d'eau par Tube !!!!!!!!!!!!!
Used quantities :
- DNA : 2µL
- Mix : (it was divided in 6 tubes for each promotor with 23µL of mix in each tube)
- Oligo 44 : 17.5µL
- Oligo 43 : 17.5µL
- Buffer Dream Taq : 87.5µL
- dNTP : 17.5µL
- Dream Taq : 7µL
- H2O : 591µL
PCR Program :
 "
"