Team:NYMU-Taipei/Project/Inhibition
From 2013.igem.org
(Created page with "{{:Team:NYMU-Taipei/Header}} {{:Team:NYMU-Taipei/Footer}}") |
Mastershot (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| + | |||
| + | =Nosema spp. indentification= | ||
| + | == Introduction == | ||
| + | |||
| + | The first step to prevent Nosema ceranae’s attack is to inhibit the spore germination occuring in a bee’s midgut. To reach our goal, the Bee. coli will continuously secrete mannosidase to destruct Nosema ceranae’s polar tube protein. The testing method is to count the spore germination rate and the polar tube protein length under a microscope field. Moreover, before conducting the wet lab procedures mentioned above, we designed new primers based on the lately released genes rather than using 16S or 18S ribosomal RNA, which is a novel way in providing species-specific signature sequences useful for Nosema spp. identification. | ||
| + | |||
| + | ==Nosema spp. identification== | ||
| + | *Morphology: Spores of N. ceranae were oval or rod shaped, varied in size with a length 3.9--5.3 μm (mean ± S.E. = 4.4 ± 0.41 μm) and a width 2.0--2.5 μm ((mean ± S.E. = 2.2 ± 0.09 μm) (N = 50) (Fig. 1), measured approximately 4.4 × 2.2 μm on fresh smears. Nosema apis are said to be slightly longer and fatter than Nosema ceranae. The number of N. ceranae's polar filament coils is between 20 and 23, rather than the more than 30 often seen in N. apis. It is difficult to identify these two species morphologically. | ||
| + | [[File:Micro1.PNG|300px|thumb|Nosema ceranae spores]] [[File:Micro2.PNG|300px|thumb|Nosema ceranae spores]] | ||
| + | *Traditional primers: Old primers are originated from small subunit ribosomal RNA genes, which are very similar, so it is difficult to design a specific primer for a species and the generated PCR fragments are too close in length. | ||
| + | '''Pimers specific for Nosema apis:''' | ||
| + | The PCR product= 401bp. | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !binding part temp. | ||
| + | |- | ||
| + | |forward: | ||
| + | | ccattgccggataagagagt | ||
| + | |unknown | ||
| + | |- | ||
| + | |reverse: || cacgcattgctgcatcattgac || unknown | ||
| + | |} | ||
| + | '''Pimers specific for Nosema ceranae:''' | ||
| + | The PCR product= 250bp. | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !binding part temp. | ||
| + | |- | ||
| + | |forward: | ||
| + | | cggataaaagagtccgttacc | ||
| + | |unknown | ||
| + | |- | ||
| + | |reverse: || tgagcagggttctagggatc || unknown | ||
| + | |} | ||
| + | '''Pimers for Nosema apis and Nosema ceranae:''' | ||
| + | The PCR product= 208bp. | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !binding part temp. | ||
| + | |- | ||
| + | |forward: | ||
| + | | ggcagttatgggaagtaaca | ||
| + | |unknown | ||
| + | |- | ||
| + | |reverse: || ggtcgtcacatttcatctct || unknown | ||
| + | |} | ||
| + | |||
| + | *New primers: As the whole genome shotgun contigs of Nosema apis and Nosema ceranae has been released lately, we chose genes which exist in only one species respectively to identify Nosema spp., which is more accurate than using SSUrRNA genes when identifying similar species. The new primers were verified by BLAST. | ||
| + | *'''nucletide sequence of class iv chitinase in N. apis:'''<br> | ||
| + | ttagtaacat ccttcttcag aagctaaatt ttctacacct | ||
| + | 29881 aaaatttctg caacatcttt atatatttta tacctatttt gacttttttc taaatttgat | ||
| + | 29941 cctgtacatt ctaatgatcc attaattgct ttagtagttg ctccaaactg attatcgcta | ||
| + | 30001 acaccaggcg cagatgcaac tttatttttc caaaaccaag cgctaacttt tgctcccaat | ||
| + | 30061 tcaggatcct ctgctacttt ttcaggatta tcaaaaatat cttctccgat tgcttcacct | ||
| + | 30121 gcttctttat aattttcagg ccatgataat tgaataaaac cacgaccgtg atatgatttt | ||
| + | 30181 ccaggagctc ctccactacc tccatattgc tcggcacatt tatttgaccc agcgcaatct | ||
| + | 30241 atttcttcta tatattgaaa tcctccagat tcatgtcctg tttgtgcaat aaacattgct | ||
| + | 30301 gcttcattca aatcattaaa ttcttcatta acaaccttga caattgcatc tacaaattct | ||
| + | 30361 ggatttggtt gataattgct tttagtcatt gcttctatta tttgatcaga agtaatattt | ||
| + | 30421 aaatttccac ctcctccact gctattgcca ccagcctttg aaccttgaga attagaagca | ||
| + | 30481 cccatagaac cagtattttt actagtgtta gcattttgtg attgtgttga agaagtacct | ||
| + | 30541 gtactttttg ttcctccttg aacattattt actggatttg aagcattatt aaaattagca | ||
| + | 30601 ttattatttg ttttcaaagc actggaacca ttggaactac cgcttgtttt tgaagcacca | ||
| + | 30661 gaagaactat ttttagaacc ccctccacca ttagttccta aatttgagat tggaccattt | ||
| + | 30721 tgtgaagatc ctgattttgc agaagaacca tttttagaac ttcctccacc actagttcct | ||
| + | 30781 aaattagaaa ttggaccact ttgtgaagat cctgattttg cagaagaacc atttttagaa | ||
| + | 30841 cctcctccac cactagtttc taaattagag attggaccac tttgtgaaga tcctgatttt | ||
| + | 30901 acagaagaac catttgacga atttttcatg ttactactat tatttgattt tgatgaattt | ||
| + | 30961 tgttgtgcaa tattagatat tggtttatta ttatttccgt tcgaacttgt accagaacca | ||
| + | 31021 atcaaatttg aacttttaac atttggtttt attttgttat cttttttctt atttatatca | ||
| + | 31081 atgtttacac caacttggtt ttcatttttt aaaatagttc gagttactgt tacagtagtt | ||
| + | 31141 attggatttt gaagttgcat<br> | ||
| + | '''Pimers specific for Nosema apis:''' | ||
| + | Nosema apis class iv chitinase only exists in N. apis and is labeled as NAPIS_ORF02138,LOCUS EQB60308 in Genebank. The PCR product= 1320bp. | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !binding part temp. | ||
| + | |- | ||
| + | |forward: | ||
| + | | ttagtaacatccttcttcagaagctaa | ||
| + | |56℃ | ||
| + | |- | ||
| + | |reverse: || atgcaacttcaaaatccaataactactg || 56℃ | ||
| + | |} | ||
| + | |||
| + | |||
| + | *'''nucletide sequence of chitin synthase d in N. apis:'''<br> | ||
| + | atgttatcac | ||
| + | 5041 aaggagaact acttaggaat ccatcaagaa ccattttaca aagaccttta aaacctcgaa | ||
| + | 5101 tcaaaaaacc aggtttatgg tcaaaattat caaggttatt tacttgtctt attccaaact | ||
| + | 5161 ttttattaag atggtgtgga atggaaacag aagaaattca gcgtgcgtgg cgggaaaaag | ||
| + | 5221 ttgctctttg tatgtgtata tttatttgtt ggattatatt aactttttta acatatggga | ||
| + | 5281 tgaatagtat cgtatgtaaa ggcaacaatc aatatattta tggacaactt cataaagcaa | ||
| + | 5341 agttcgaaag aaatatagtt attacaaatg gaagtatatt ttacacagac gatgattcat | ||
| + | 5401 atgaaccaaa tcaaatttat actggaactt ttgaaaataa aagttctgca tgtaaaaaag | ||
| + | 5461 catttggacg acaattatta agtggaggaa gaagtactaa aaaattagaa agaattgcac | ||
| + | 5521 caatttgttt tgattgggca tttattacta aaaataattt tattacgatc ggaaataagg | ||
| + | 5581 tttatgatac ttccttatgc aaagaagaaa cttatgataa ttttataaaa aaatactctg | ||
| + | 5641 gaaccgaagc taaacaagat tatttaacag atgatgaatg gcaatgtttt aaagacgcat | ||
| + | 5701 ttttttccgg agaaattgct acaaaaacac ctggatgctt tatagcagat acttttttgt | ||
| + | 5761 atattacaac cgtaataatt ttttcattga ttattgcaaa atttgtactt gcaactgttt | ||
| + | 5821 attcttggca tatgagaaga aaagtacgtc ctactgaaca gataacgcct tgtatattac | ||
| + | 5881 ttgttacagc atatagcgaa gatgaagaag gtttgagaaa aactcttgat agtctttgca | ||
| + | 5941 ctcaacaata taattatgat agaaaactaa tcattattat ttctgatgga gaaataacag | ||
| + | 6001 gagaagaaaa tgcaaaaagt acaccagata ttattttaga tttaatggaa attgatgata | ||
| + | 6061 gtgtagttga accaaaaagc tatgtatctt taatgcctgg ttcgaaacga ataaataaag | ||
| + | 6121 caaaagtata tacaggatac tataattcta gagacagatc tagtaattat aataaatgtc | ||
| + | 6181 gaatattatt tataagaaaa tgtggaaatt cttcagaatc atttaaagca ggtaatagag | ||
| + | 6241 gaaagagaga cactcaagta attttaatgt catttttttc caaactaata tatggtgata | ||
| + | 6301 gaatgtcaga acttgatttt gaactttacc ttcgtttaaa aaaattaatg ccagaagtgg | ||
| + | 6361 aacctgtaga ttttgaatgt atgcttatgg tagatgcaga taccatcgta aaacccgatg | ||
| + | 6421 ctctaagtaa aatggtatcg atatttgaaa cggatccaaa agttatagga atttgtggag | ||
| + | 6481 aaacaatgat tttaaataaa tgtgaaagtt gggtatcaat gatacaggtc tttgagtatt | ||
| + | 6541 atataagtca tcatttaagc aaaacatttg aatcagtctt cggaggttta acttgtcttc | ||
| + | 6601 caggatgttt ttgtatgtat agaatcaaaa taattacaga cgaagatgga aatgttatta | ||
| + | 6661 ctggatttaa gtcggtaaat tcaaaaattt ttagtgatga ttcttatgaa aaaaactgct | ||
| + | 6721 tacctatttt ggcaaatcca tttataataa attcatattc tgtatttgaa gccaaaacac | ||
| + | 6781 ttcatcaaaa aaatttactg catttaggag aagatagata tttaacaacg cttcttctta | ||
| + | 6841 aaaatttcta caaaagaaaa ttaatttttg tgccgcaagc aaaatgcgaa acatatgttc | ||
| + | 6901 ctagtaaatt taaagtatta ttaagtcaaa ggagaagatg gataaactca acaatacaca | ||
| + | 6961 atttacttga attaatgatg gtagataaat tgtgcggagt attttgttgt tcaatgcaat | ||
| + | 7021 ttgttgttat tgttgaatta tttggaactt tagtacttcc agctgcaata cttttcacag | ||
| + | 7081 gtgttttgat agttacttca attttatacg aacctgcatg gataccactt ataatgttat | ||
| + | 7141 ttgggatttt tattttaccg gcttttctaa tcttgattac gacattcgaa atcagttata | ||
| + | 7201 tattctggct ttttatatat attctatcaa ttcctatatg gaattttgta ttaccaatat | ||
| + | 7261 atgcattttg gcattttgat gatttttcat ggggagatac gaggaaagta acaggtgaaa | ||
| + | 7321 ataaggaaga aataaataaa aatgacgaac aagaaaaaat aaaattaatg gagttagaag | ||
| + | 7381 attatctaca agaaattaat gaatag | ||
| + | <br> | ||
| + | '''Pimers for Nosema apis and Nosema ceranae:''' | ||
| + | Nosema chitin synthase d is labeled as NAPIS_ORF01775 in Genebank. A very similar sequence is found in Nosema ceranae BRL01 Nc000006, whole genome shotgun sequence, where the Sequence ID: gb|ACOL01000006.1|. The PCR product=1089bp for N.apis and 1103bp for N.ceranae. | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !binding part temp. | ||
| + | |- | ||
| + | |forward: | ||
| + | | AATTTATACTGGAACTTTTGAAAATAAAAGT | ||
| + | |54℃ | ||
| + | |- | ||
| + | |reverse: || CATTTATTTAAAATCATTGTTTCTCCACAAAT || 55℃ | ||
| + | |} | ||
| + | |||
| + | |||
| + | =Polar Filament Inhibition= | ||
| + | ==background== | ||
| + | After ''N. ceranae'' invades the epithelial cell of bee's midgut it will soon reproduce in two days and new generation appears. Since it's lifecycle is really short that there are no time for biological methods to react and inhibit the pathogen's development, we choose to inhibit it before it enter's the bee's epithelial cell, which will be before ''N. ceranae''’s germination. When ''N. ceranae'''s spore moves to bee's midgut it will start to germinate and develop a structure called polar filament. This structure will then bind to the epithelial cell of the bee's midgut and poke a hole at the membrane of epithelial cell, causing the bee eventually dead.In order to inhibit polar filament's development, we have found out that there is a protein called PTP1 which is highly relevant to the binding of the polar filament and the epithelial cell. So we decided to suppress the growth of the polar filament by obstruct the production of PTP1. In the research that is done by other scientist, they have found out that in the process of PTP1 formation a mannose will bind to the protein. So we use mannosidase to prevent ''N. ceranae'' from producing proper PTP1 further to stop its polar filament to bind to the epithelial cell and of course, stop the infection of ''N. ceranae''. | ||
| + | ==mannosidase== | ||
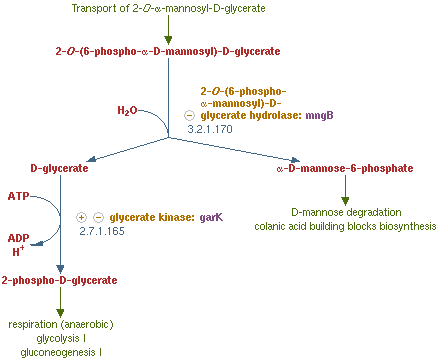
| + | [[File:NYMU_Taipei_MngB pathway.PNG|right|thumb|325px|2-O-α-mannosyl-D-glycerate degradation pathway in MG1655]] | ||
| + | Mannosidase is an enzyme that can be found in many different organisms. It is an enzyme that turns 2-O-(6-phospho-α-mannosyl)-D-glycerate into mannose-6-phosphate and glycerate, which is the reverse reaction of the reaction that mentioned before in the PTP1's formation. This reaction is part of the 2-O-α-mannosyl-D-glycerate degradation pathway, which is often seen in many organisms.We choose to clone the mannosidase gene that is from Escherichia coli K-12 substr. MG1655 which is also the bacteria that we are going to transform our parts into and form our devise. This can let our experiment become more easier and be more sure that the enzyme can have the right function in our Bee. coli. This gene is called mngB. We overexpress this gene in order to inhibit the production of the PTP1 in the polar filament of ''N. ceranae''.<br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==circuit design== | ||
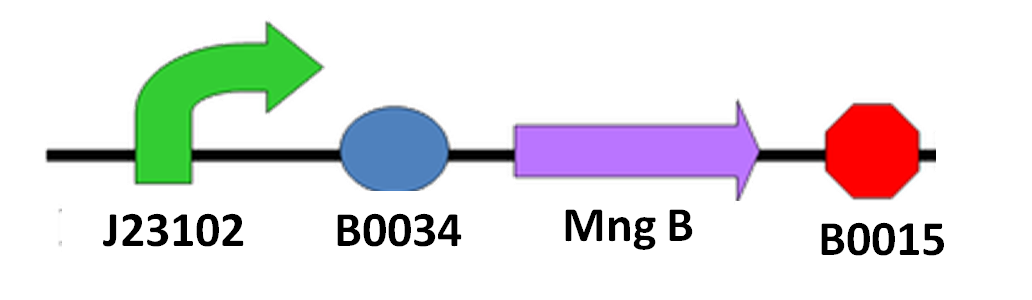
| + | Since we cannot be sure when the spore of ''N. ceranae'' will start growing polar filament, also not having an accurate sensor to know when ''N. ceranae'' invade, we decided to add a constitutive promoter in front of mngB to let it express continuously. <br>The circuit design is as below:<br> | ||
| + | [[File:NYMU_Taipei_mannosidase circuit.PNG|left|thumb|350px|circuit design]] | ||
| + | |||
| + | ==experiment== | ||
| + | |||
| + | '''alpha mannosidase primer:''' | ||
| + | {|class="wikitable" | ||
| + | |- | ||
| + | ! | ||
| + | !primer sequence | ||
| + | !whole primer temp. | ||
| + | !binding part temp. | ||
| + | !GC% | ||
| + | |- | ||
| + | |forward: | ||
| + | | ctg GAATTCGCGGCCGCTTCTAG atgAAAGCAGTATCTCGCGTTCACATCACCCCG | ||
| + | |76℃ | ||
| + | |68℃ ||55% | ||
| + | |- | ||
| + | |reverse: || gga CTGCAGCGGCCGCTACTAGTA tcaGGCAAGCCGGTAACTGAACGTCCG ||76℃ ||68℃ ||58% | ||
| + | |} | ||
| + | |||
| + | ==reference== | ||
| + | link:[http://ecocyc.org/ECOLI/NEW-IMAGE?type=ENZYME&object=EG13236-MONOMER alpha mannosidase] | ||
| + | |||
| + | =protocol= | ||
| + | ==DNA extraction== | ||
| + | Use the same kit as bacterial genome extraction. | ||
| + | ==Germination method== | ||
| + | 1. Take 1cc N. ceranae mixture in an eppendorf and then centrifuge at 13200 rpm for 20 minutes. | ||
| + | |||
| + | 2. Discard the supernatant and then pipeting the pellet with 0.1M sucrose in PBS until the pellet is dissolved. | ||
| + | |||
| + | 3. Put the eppendorf at 37 Celsius Degree in the incubator for 12 hours. | ||
| + | ==Testing method== | ||
| + | 1. Take 5λ liquid to make the glass slide for N. ceranae sample. | ||
| + | |||
| + | 2. Manipulate the light microscope and take photos. | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Revision as of 10:56, 23 September 2013


Contents |
Nosema spp. indentification
Introduction
The first step to prevent Nosema ceranae’s attack is to inhibit the spore germination occuring in a bee’s midgut. To reach our goal, the Bee. coli will continuously secrete mannosidase to destruct Nosema ceranae’s polar tube protein. The testing method is to count the spore germination rate and the polar tube protein length under a microscope field. Moreover, before conducting the wet lab procedures mentioned above, we designed new primers based on the lately released genes rather than using 16S or 18S ribosomal RNA, which is a novel way in providing species-specific signature sequences useful for Nosema spp. identification.
Nosema spp. identification
- Morphology: Spores of N. ceranae were oval or rod shaped, varied in size with a length 3.9--5.3 μm (mean ± S.E. = 4.4 ± 0.41 μm) and a width 2.0--2.5 μm ((mean ± S.E. = 2.2 ± 0.09 μm) (N = 50) (Fig. 1), measured approximately 4.4 × 2.2 μm on fresh smears. Nosema apis are said to be slightly longer and fatter than Nosema ceranae. The number of N. ceranae's polar filament coils is between 20 and 23, rather than the more than 30 often seen in N. apis. It is difficult to identify these two species morphologically.
- Traditional primers: Old primers are originated from small subunit ribosomal RNA genes, which are very similar, so it is difficult to design a specific primer for a species and the generated PCR fragments are too close in length.
Pimers specific for Nosema apis: The PCR product= 401bp.
| primer sequence | binding part temp. | |
|---|---|---|
| forward: | ccattgccggataagagagt | unknown |
| reverse: | cacgcattgctgcatcattgac | unknown |
Pimers specific for Nosema ceranae: The PCR product= 250bp.
| primer sequence | binding part temp. | |
|---|---|---|
| forward: | cggataaaagagtccgttacc | unknown |
| reverse: | tgagcagggttctagggatc | unknown |
Pimers for Nosema apis and Nosema ceranae: The PCR product= 208bp.
| primer sequence | binding part temp. | |
|---|---|---|
| forward: | ggcagttatgggaagtaaca | unknown |
| reverse: | ggtcgtcacatttcatctct | unknown |
- New primers: As the whole genome shotgun contigs of Nosema apis and Nosema ceranae has been released lately, we chose genes which exist in only one species respectively to identify Nosema spp., which is more accurate than using SSUrRNA genes when identifying similar species. The new primers were verified by BLAST.
- nucletide sequence of class iv chitinase in N. apis:
ttagtaacat ccttcttcag aagctaaatt ttctacacct 29881 aaaatttctg caacatcttt atatatttta tacctatttt gacttttttc taaatttgat 29941 cctgtacatt ctaatgatcc attaattgct ttagtagttg ctccaaactg attatcgcta 30001 acaccaggcg cagatgcaac tttatttttc caaaaccaag cgctaacttt tgctcccaat 30061 tcaggatcct ctgctacttt ttcaggatta tcaaaaatat cttctccgat tgcttcacct 30121 gcttctttat aattttcagg ccatgataat tgaataaaac cacgaccgtg atatgatttt 30181 ccaggagctc ctccactacc tccatattgc tcggcacatt tatttgaccc agcgcaatct 30241 atttcttcta tatattgaaa tcctccagat tcatgtcctg tttgtgcaat aaacattgct 30301 gcttcattca aatcattaaa ttcttcatta acaaccttga caattgcatc tacaaattct 30361 ggatttggtt gataattgct tttagtcatt gcttctatta tttgatcaga agtaatattt 30421 aaatttccac ctcctccact gctattgcca ccagcctttg aaccttgaga attagaagca 30481 cccatagaac cagtattttt actagtgtta gcattttgtg attgtgttga agaagtacct 30541 gtactttttg ttcctccttg aacattattt actggatttg aagcattatt aaaattagca 30601 ttattatttg ttttcaaagc actggaacca ttggaactac cgcttgtttt tgaagcacca 30661 gaagaactat ttttagaacc ccctccacca ttagttccta aatttgagat tggaccattt 30721 tgtgaagatc ctgattttgc agaagaacca tttttagaac ttcctccacc actagttcct 30781 aaattagaaa ttggaccact ttgtgaagat cctgattttg cagaagaacc atttttagaa 30841 cctcctccac cactagtttc taaattagag attggaccac tttgtgaaga tcctgatttt 30901 acagaagaac catttgacga atttttcatg ttactactat tatttgattt tgatgaattt 30961 tgttgtgcaa tattagatat tggtttatta ttatttccgt tcgaacttgt accagaacca 31021 atcaaatttg aacttttaac atttggtttt attttgttat cttttttctt atttatatca 31081 atgtttacac caacttggtt ttcatttttt aaaatagttc gagttactgt tacagtagtt 31141 attggatttt gaagttgcat
Pimers specific for Nosema apis: Nosema apis class iv chitinase only exists in N. apis and is labeled as NAPIS_ORF02138,LOCUS EQB60308 in Genebank. The PCR product= 1320bp.
| primer sequence | binding part temp. | |
|---|---|---|
| forward: | ttagtaacatccttcttcagaagctaa | 56℃ |
| reverse: | atgcaacttcaaaatccaataactactg | 56℃ |
- nucletide sequence of chitin synthase d in N. apis:
atgttatcac
5041 aaggagaact acttaggaat ccatcaagaa ccattttaca aagaccttta aaacctcgaa
5101 tcaaaaaacc aggtttatgg tcaaaattat caaggttatt tacttgtctt attccaaact
5161 ttttattaag atggtgtgga atggaaacag aagaaattca gcgtgcgtgg cgggaaaaag
5221 ttgctctttg tatgtgtata tttatttgtt ggattatatt aactttttta acatatggga
5281 tgaatagtat cgtatgtaaa ggcaacaatc aatatattta tggacaactt cataaagcaa
5341 agttcgaaag aaatatagtt attacaaatg gaagtatatt ttacacagac gatgattcat
5401 atgaaccaaa tcaaatttat actggaactt ttgaaaataa aagttctgca tgtaaaaaag
5461 catttggacg acaattatta agtggaggaa gaagtactaa aaaattagaa agaattgcac
5521 caatttgttt tgattgggca tttattacta aaaataattt tattacgatc ggaaataagg
5581 tttatgatac ttccttatgc aaagaagaaa cttatgataa ttttataaaa aaatactctg
5641 gaaccgaagc taaacaagat tatttaacag atgatgaatg gcaatgtttt aaagacgcat
5701 ttttttccgg agaaattgct acaaaaacac ctggatgctt tatagcagat acttttttgt
5761 atattacaac cgtaataatt ttttcattga ttattgcaaa atttgtactt gcaactgttt
5821 attcttggca tatgagaaga aaagtacgtc ctactgaaca gataacgcct tgtatattac
5881 ttgttacagc atatagcgaa gatgaagaag gtttgagaaa aactcttgat agtctttgca
5941 ctcaacaata taattatgat agaaaactaa tcattattat ttctgatgga gaaataacag
6001 gagaagaaaa tgcaaaaagt acaccagata ttattttaga tttaatggaa attgatgata
6061 gtgtagttga accaaaaagc tatgtatctt taatgcctgg ttcgaaacga ataaataaag
6121 caaaagtata tacaggatac tataattcta gagacagatc tagtaattat aataaatgtc
6181 gaatattatt tataagaaaa tgtggaaatt cttcagaatc atttaaagca ggtaatagag
6241 gaaagagaga cactcaagta attttaatgt catttttttc caaactaata tatggtgata
6301 gaatgtcaga acttgatttt gaactttacc ttcgtttaaa aaaattaatg ccagaagtgg
6361 aacctgtaga ttttgaatgt atgcttatgg tagatgcaga taccatcgta aaacccgatg
6421 ctctaagtaa aatggtatcg atatttgaaa cggatccaaa agttatagga atttgtggag
6481 aaacaatgat tttaaataaa tgtgaaagtt gggtatcaat gatacaggtc tttgagtatt
6541 atataagtca tcatttaagc aaaacatttg aatcagtctt cggaggttta acttgtcttc
6601 caggatgttt ttgtatgtat agaatcaaaa taattacaga cgaagatgga aatgttatta
6661 ctggatttaa gtcggtaaat tcaaaaattt ttagtgatga ttcttatgaa aaaaactgct
6721 tacctatttt ggcaaatcca tttataataa attcatattc tgtatttgaa gccaaaacac
6781 ttcatcaaaa aaatttactg catttaggag aagatagata tttaacaacg cttcttctta
6841 aaaatttcta caaaagaaaa ttaatttttg tgccgcaagc aaaatgcgaa acatatgttc
6901 ctagtaaatt taaagtatta ttaagtcaaa ggagaagatg gataaactca acaatacaca
6961 atttacttga attaatgatg gtagataaat tgtgcggagt attttgttgt tcaatgcaat
7021 ttgttgttat tgttgaatta tttggaactt tagtacttcc agctgcaata cttttcacag
7081 gtgttttgat agttacttca attttatacg aacctgcatg gataccactt ataatgttat
7141 ttgggatttt tattttaccg gcttttctaa tcttgattac gacattcgaa atcagttata
7201 tattctggct ttttatatat attctatcaa ttcctatatg gaattttgta ttaccaatat
7261 atgcattttg gcattttgat gatttttcat ggggagatac gaggaaagta acaggtgaaa
7321 ataaggaaga aataaataaa aatgacgaac aagaaaaaat aaaattaatg gagttagaag
7381 attatctaca agaaattaat gaatag
Pimers for Nosema apis and Nosema ceranae:
Nosema chitin synthase d is labeled as NAPIS_ORF01775 in Genebank. A very similar sequence is found in Nosema ceranae BRL01 Nc000006, whole genome shotgun sequence, where the Sequence ID: gb|ACOL01000006.1|. The PCR product=1089bp for N.apis and 1103bp for N.ceranae.
| primer sequence | binding part temp. | |
|---|---|---|
| forward: | AATTTATACTGGAACTTTTGAAAATAAAAGT | 54℃ |
| reverse: | CATTTATTTAAAATCATTGTTTCTCCACAAAT | 55℃ |
Polar Filament Inhibition
background
After N. ceranae invades the epithelial cell of bee's midgut it will soon reproduce in two days and new generation appears. Since it's lifecycle is really short that there are no time for biological methods to react and inhibit the pathogen's development, we choose to inhibit it before it enter's the bee's epithelial cell, which will be before N. ceranae’s germination. When N. ceranae's spore moves to bee's midgut it will start to germinate and develop a structure called polar filament. This structure will then bind to the epithelial cell of the bee's midgut and poke a hole at the membrane of epithelial cell, causing the bee eventually dead.In order to inhibit polar filament's development, we have found out that there is a protein called PTP1 which is highly relevant to the binding of the polar filament and the epithelial cell. So we decided to suppress the growth of the polar filament by obstruct the production of PTP1. In the research that is done by other scientist, they have found out that in the process of PTP1 formation a mannose will bind to the protein. So we use mannosidase to prevent N. ceranae from producing proper PTP1 further to stop its polar filament to bind to the epithelial cell and of course, stop the infection of N. ceranae.
mannosidase
Mannosidase is an enzyme that can be found in many different organisms. It is an enzyme that turns 2-O-(6-phospho-α-mannosyl)-D-glycerate into mannose-6-phosphate and glycerate, which is the reverse reaction of the reaction that mentioned before in the PTP1's formation. This reaction is part of the 2-O-α-mannosyl-D-glycerate degradation pathway, which is often seen in many organisms.We choose to clone the mannosidase gene that is from Escherichia coli K-12 substr. MG1655 which is also the bacteria that we are going to transform our parts into and form our devise. This can let our experiment become more easier and be more sure that the enzyme can have the right function in our Bee. coli. This gene is called mngB. We overexpress this gene in order to inhibit the production of the PTP1 in the polar filament of N. ceranae.
circuit design
Since we cannot be sure when the spore of N. ceranae will start growing polar filament, also not having an accurate sensor to know when N. ceranae invade, we decided to add a constitutive promoter in front of mngB to let it express continuously.
The circuit design is as below:
experiment
alpha mannosidase primer:
| primer sequence | whole primer temp. | binding part temp. | GC% | |
|---|---|---|---|---|
| forward: | ctg GAATTCGCGGCCGCTTCTAG atgAAAGCAGTATCTCGCGTTCACATCACCCCG | 76℃ | 68℃ | 55% |
| reverse: | gga CTGCAGCGGCCGCTACTAGTA tcaGGCAAGCCGGTAACTGAACGTCCG | 76℃ | 68℃ | 58% |
reference
link:[http://ecocyc.org/ECOLI/NEW-IMAGE?type=ENZYME&object=EG13236-MONOMER alpha mannosidase]
protocol
DNA extraction
Use the same kit as bacterial genome extraction.
Germination method
1. Take 1cc N. ceranae mixture in an eppendorf and then centrifuge at 13200 rpm for 20 minutes.
2. Discard the supernatant and then pipeting the pellet with 0.1M sucrose in PBS until the pellet is dissolved.
3. Put the eppendorf at 37 Celsius Degree in the incubator for 12 hours.
Testing method
1. Take 5λ liquid to make the glass slide for N. ceranae sample.
2. Manipulate the light microscope and take photos.
 "
"