Team:British Columbia/Project/CRISPR
From 2013.igem.org
(→CRISPR) |
|||
| Line 10: | Line 10: | ||
===[[Team:British_Columbia/Project/CRISPR/SpacerSelection|Spacer Selection]]=== | ===[[Team:British_Columbia/Project/CRISPR/SpacerSelection|Spacer Selection]]=== | ||
| + | |||
| + | ===Design=== | ||
| + | |||
| + | ===Results=== | ||
| + | |||
| + | ====Normal growth under T4 Phage Infection==== | ||
| + | |||
| + | ====Combinatorial CRISPR-Cas9 element assembly==== | ||
| + | [[File:UBC-CRISPR-Screen-Scatter.png|800px]] | ||
| + | |||
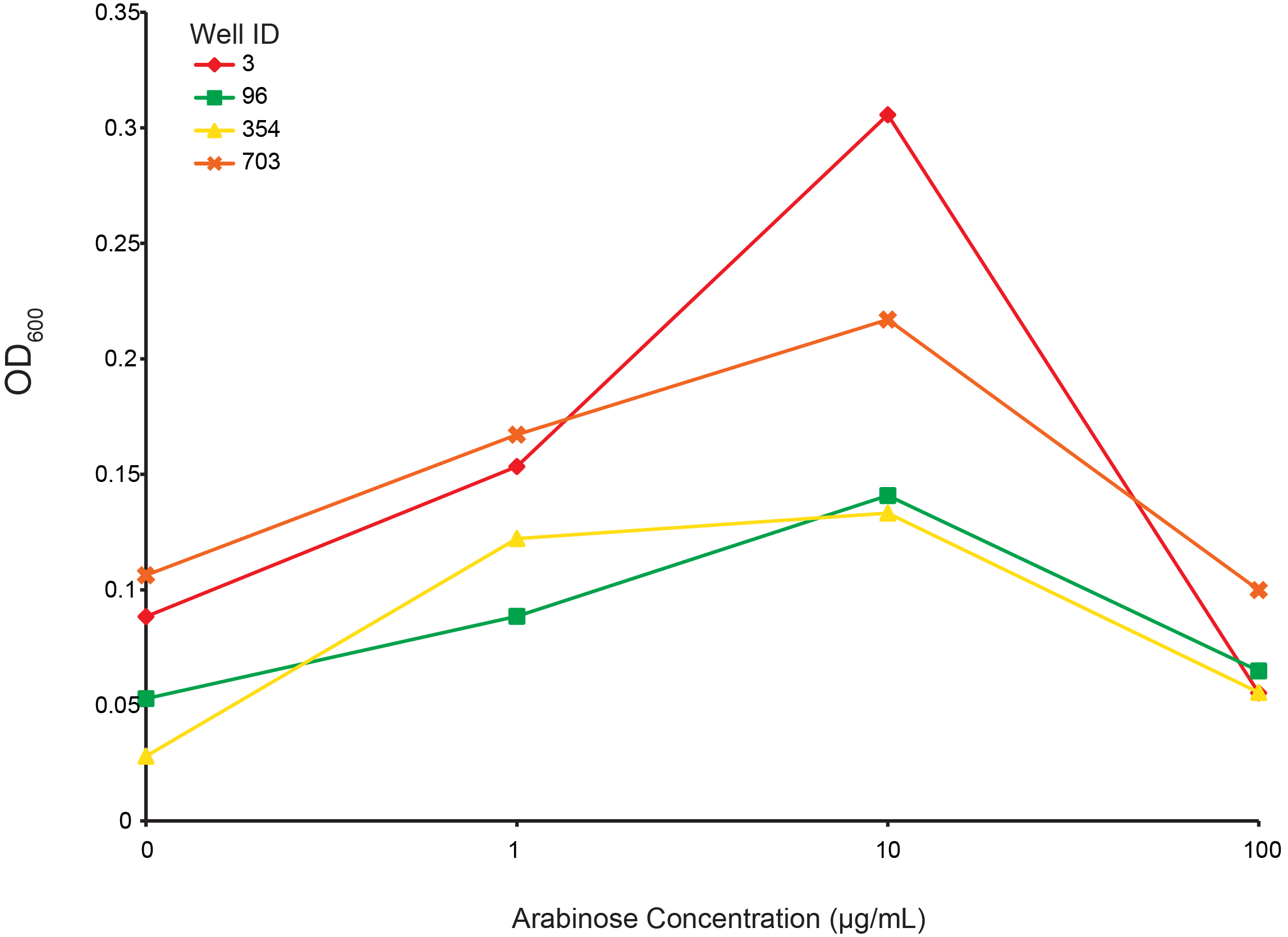
| + | ====Arabinose dependent growth under T4 infection==== | ||
| + | [[File:UBC-CRISPR-induction-dependence.png|800px]] | ||
| + | |||
| + | ====Growth kinetics of immunized culture==== | ||
| + | [[File:UBC-CRISPR-Growth.png|800px]] | ||
Revision as of 23:49, 25 September 2013
iGEM Home


Contents |
CRISPR
Molecular biology has long exploited phage machinery as tools for manipulating genomes. Recently, the literature has exploded with work on the CRISPR adaptive immune system of prokaryotes, which involves nucleases that can be guided by small RNAs. The system includes loci composed of a series of repeats separated by ‘spacer’ elements that match a target sequence. This region is transcribed into the small RNAs that specify the target sequence (or protospacer) to be cleaved by a CRISPR-associated gene, Cas9. Cas9 is a double stranded DNA endonuclease with single nucleotide resolution. Several groups have demonstrated that the CRISPR system is extremely useful for genome editing, which inspired us to re-factor it for our own purposes.
As an adaptive immune response, we wanted to know if CRISPR could be put together in a modular way to confer resistance to known phage genomes - vaccinating the host. We first wrote programs capable of predicting the most broadly neutralizing spacer region from a file of compiled phage genomes. We then assembled the minimum components biobricks and conducted the necessary proof-of-concept experiments in E.coli. First, we characterized the dynamics of phage infection in our specific host strain and experimental protocols. We then built a system that protects E.coli against T4 phage infection and are beginning to understand some guidelines for assembling CRISPR components. Currently, we are carrying out experiments with T7 phage, performing some in vitro characterizations, and exploring new possibilities with our working and manipulable CRISPR system.
Ultimately, we hope that large-scale fermenters could be vaccinated against collapse caused by environmental phage infection. To extend the application of this approach, we designed specifically neutralization elements that allow for population level programming of a culture. Check out our population control section where we envision this being first applied to yogurt where, for example, the biosynthesis of flavour combinations is controlled by targeted phage addition.
 "
"