Team:Leeds/Project
From 2013.igem.org
(Difference between revisions)
m |
|||
| Line 71: | Line 71: | ||
<br> | <br> | ||
==Phase I== | ==Phase I== | ||
| - | We split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within and without iGEM. The first of these looks at developing, characterising and producing basic | + | We split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within and without iGEM. The first of these looks at developing, characterising and producing basic Devices. |
| - | === | + | ===Device 1=== |
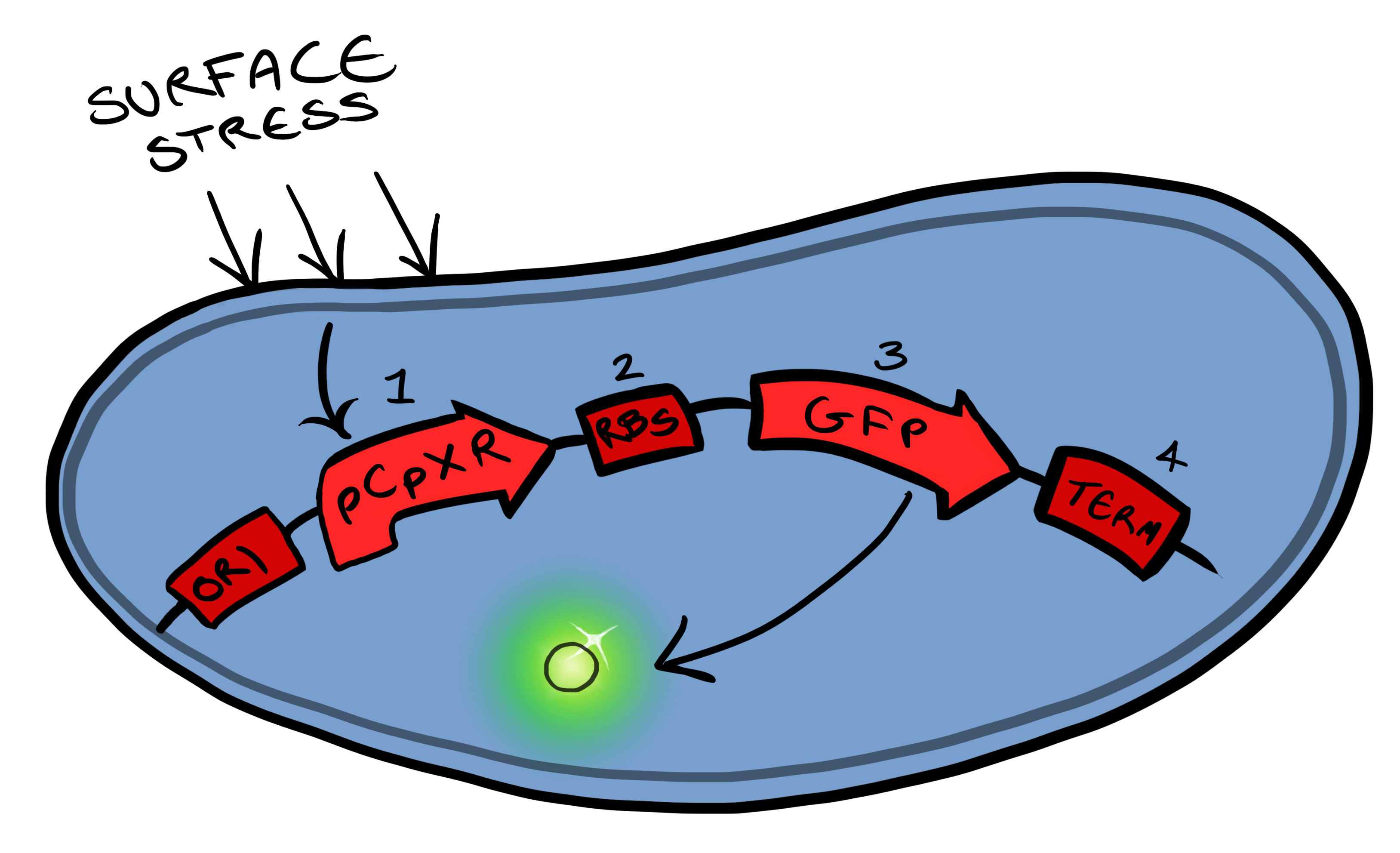
[[File:Leeds_bb1schematic.png|left|400px|Cartoon schematic of Biosystem 1|link=|frameless]] | [[File:Leeds_bb1schematic.png|left|400px|Cartoon schematic of Biosystem 1|link=|frameless]] | ||
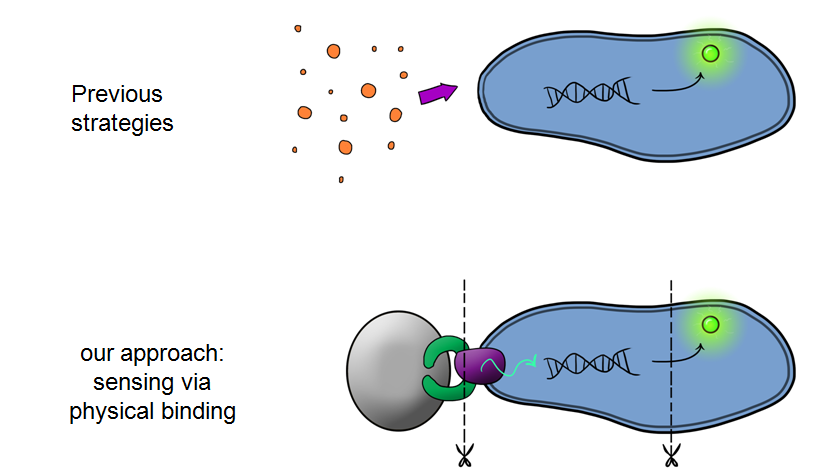
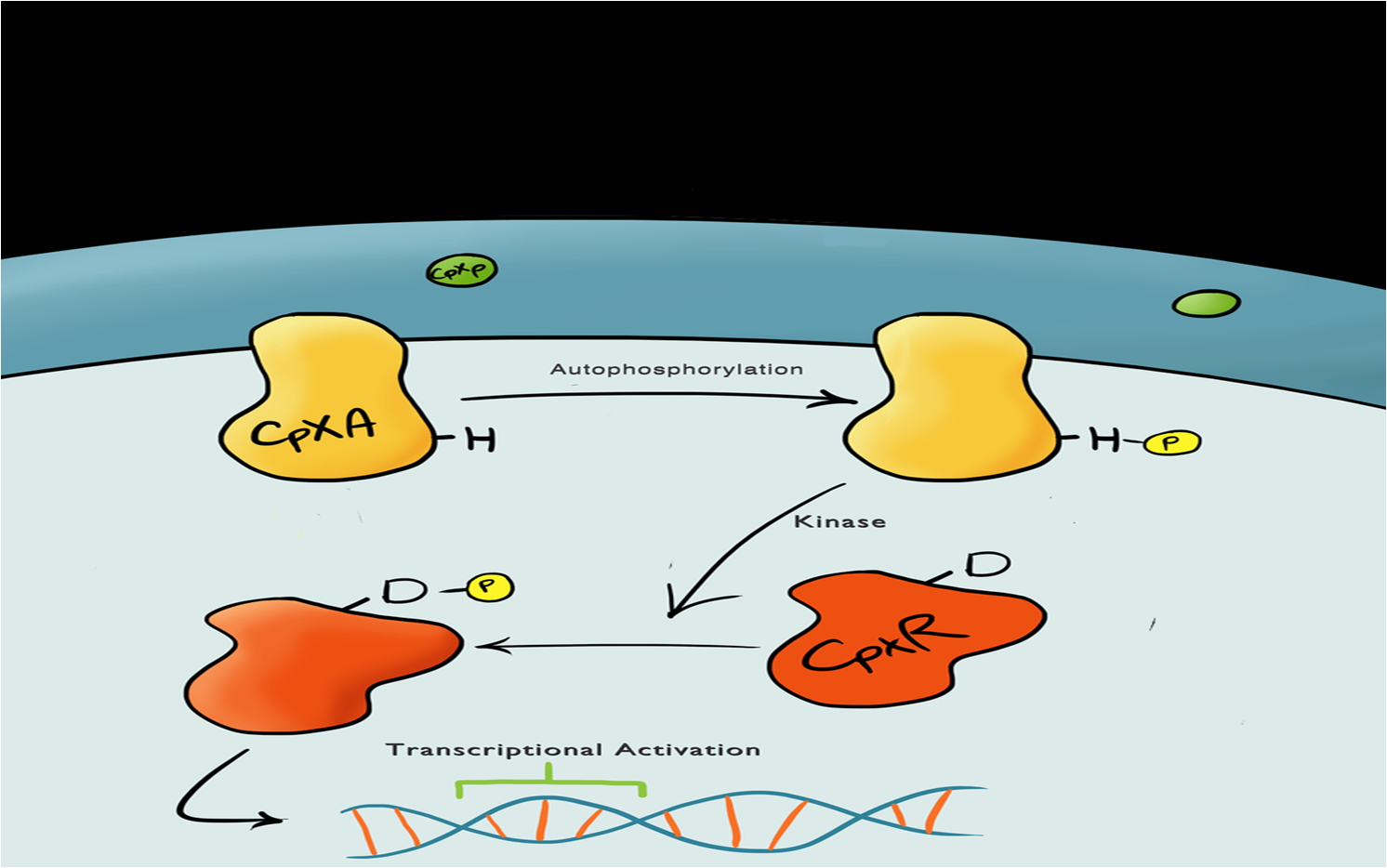
| - | + | Device 1 is a very simple modification of the Cpx promoter in ''E. coli'' to produce a fluorescent reporter when the cell is under membrane stress. We will then characterise and test the insertion following the work by [http://www.ncbi.nlm.nih.gov/pubmed/11830644 ''Otto & Silhaevy 2001''] in which glass beads were used to induce hydrophobic membrane stress. What we would hope is that when we add our cells to a vial containing similar hydrophobic beads we can detect fluorescence emissions confirming that our biobrick is working as we had hoped. We know that membrane stress can be caused by many stimuli, so we have also looked at the effect different membrane stresses cause on the florescence achieved; these membrane stresses include, pH, temperture and detergent concentration. | |
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | Bio bricks we used to assemble | + | Bio bricks we used to assemble Device 1 are: |
BBa_K135000 pCpxR promoter responds to membrane stress | BBa_K135000 pCpxR promoter responds to membrane stress | ||
BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | === | + | ===Device 2=== |
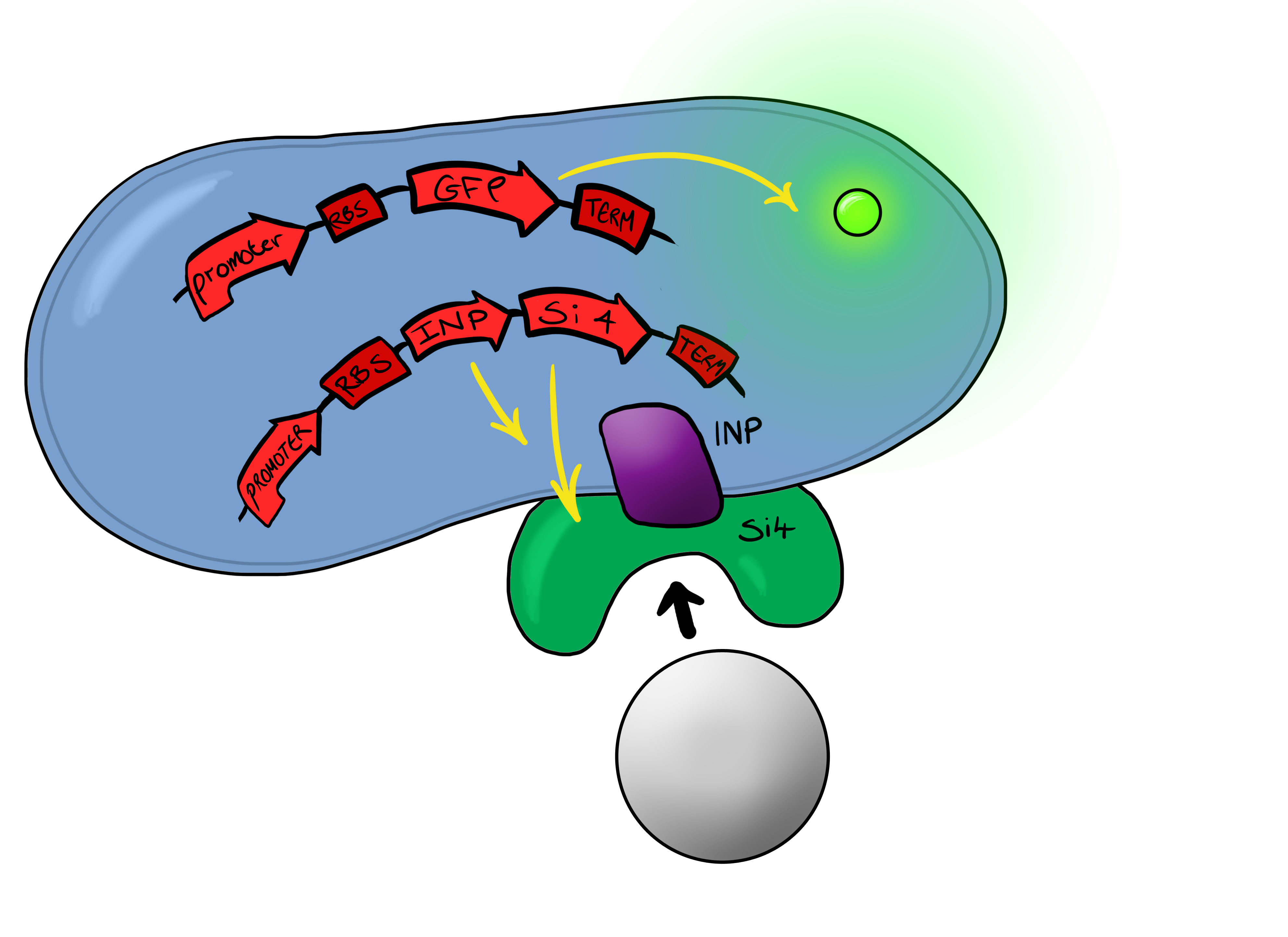
[[File:Leeds_Device2schematic.png|right|400px|Cartoon schematic of Biosystem 2|link=|frameless]] | [[File:Leeds_Device2schematic.png|right|400px|Cartoon schematic of Biosystem 2|link=|frameless]] | ||
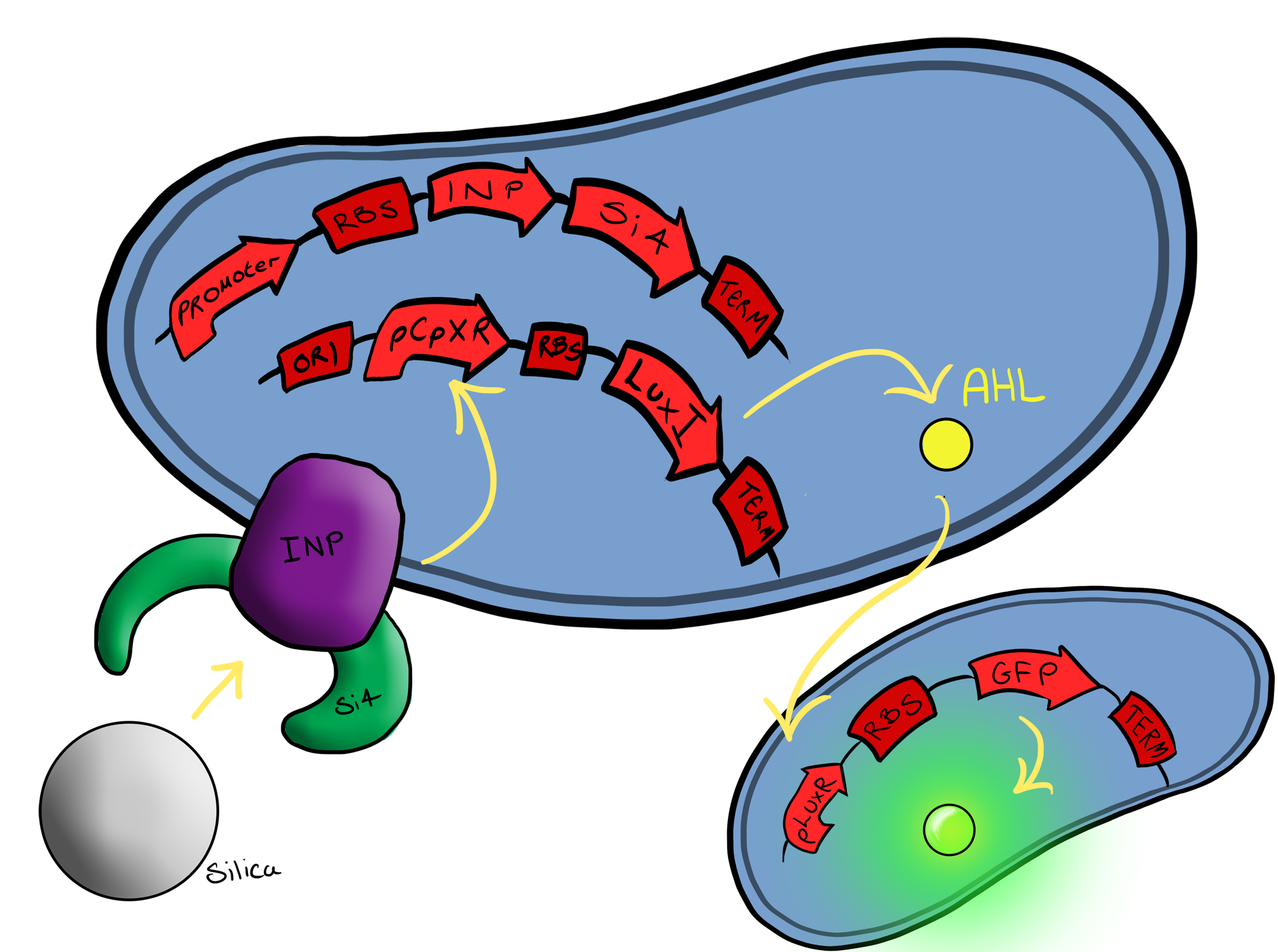
Simultaneously, we will be developing a Biosystem that is suited for physical attachment and detection of particles. This will work using Ice Nucleation Protein (INP) to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of pathogen detection. INP is a transmembrane protein that expresses any sequence that is placed on the C-terminus of its gene on the outer surface of the cell. | Simultaneously, we will be developing a Biosystem that is suited for physical attachment and detection of particles. This will work using Ice Nucleation Protein (INP) to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of pathogen detection. INP is a transmembrane protein that expresses any sequence that is placed on the C-terminus of its gene on the outer surface of the cell. | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | The cells producing this receptor complex will also be constitutively producing a fluorescent reporter, GFP, to allow for its detection during the characterisation experiments but this time the focus of | + | The cells producing this receptor complex will also be constitutively producing a fluorescent reporter, GFP, to allow for its detection during the characterisation experiments but this time the focus of Device 2 is on the adhesion proteins being used. |
<br style="clear:both"/> | <br style="clear:both"/> | ||
The Biobricks we used for this Biosystem are: | The Biobricks we used for this Biosystem are: | ||
| Line 95: | Line 95: | ||
==Phase II== | ==Phase II== | ||
Phase II takes the products of Phase I to the next level, and begins to look at integration of the two BioBricks. | Phase II takes the products of Phase I to the next level, and begins to look at integration of the two BioBricks. | ||
| - | === | + | ===Device 3=== |
[[File:Biosystem3.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | [[File:Biosystem3.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | ||
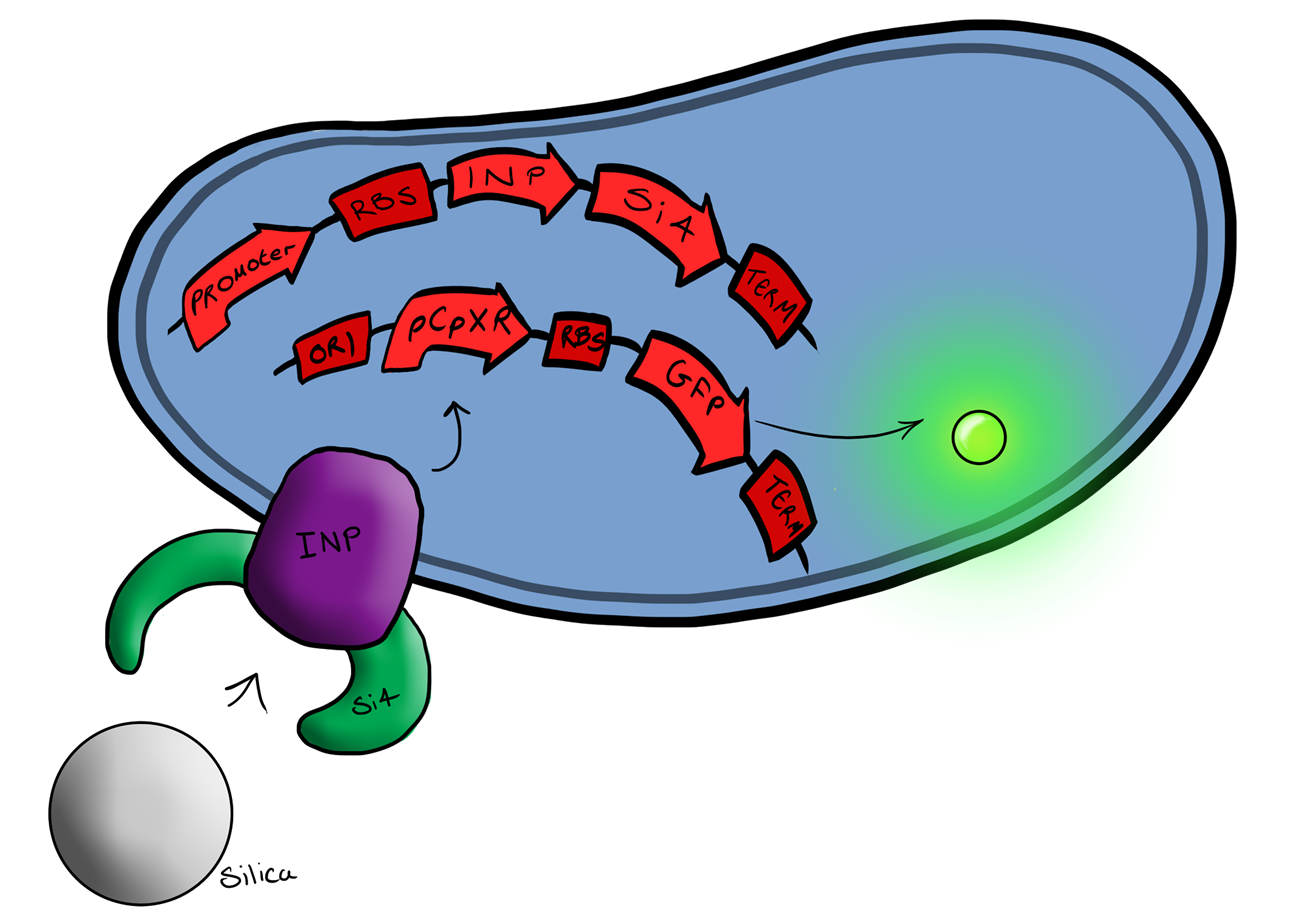
| - | + | Device 3 brings together BS1 and BS2 in one plasmid; the genes that code for the INP and the Si4 binding domain are now located on the same plasmid as the CpxP promoter and the GFP generator. The effect of this is that when the silca bead binds to the Si4 binding domain connected to the INP protein and causes surface stress the CpxP promoter will be induced. This will cause GFP to be produced and this will act as a visible signal that the 'pathogen' has been detected. | |
<br style="clear:both"/> | <br style="clear:both"/> | ||
This is the final MircoBeagle device and has the potential to be made into many different pathogen detectors. This can easily be done by swapping out the gene coding for the binding moiety next to the INP gene. Once Biosystem 3 has been suitably characterised and all bugs fixed, the next step will be to swap the silca binding domain for a binding domain that binds to a particular pathogen. | This is the final MircoBeagle device and has the potential to be made into many different pathogen detectors. This can easily be done by swapping out the gene coding for the binding moiety next to the INP gene. Once Biosystem 3 has been suitably characterised and all bugs fixed, the next step will be to swap the silca binding domain for a binding domain that binds to a particular pathogen. | ||
Revision as of 09:24, 1 October 2013
 "
"