Team:TU-Munich/HumanPractice/Education
From 2013.igem.org
(→References) |
(→References) |
||
| Line 115: | Line 115: | ||
<!-- Kopiervorlagen --> | <!-- Kopiervorlagen --> | ||
[[http://www.ncbi.nlm.nih.gov/pubmed/22368493 Close D et al., 2012]] Close D, Xu T, Smartt A, Rogers A, Crossley R, Price S, Ripp S, Sayler G (2012). The evolution of the bacterial luciferase gene cassette (lux) as a real-time bioreporter. ''Sensors'', 12(1):732-52. | [[http://www.ncbi.nlm.nih.gov/pubmed/22368493 Close D et al., 2012]] Close D, Xu T, Smartt A, Rogers A, Crossley R, Price S, Ripp S, Sayler G (2012). The evolution of the bacterial luciferase gene cassette (lux) as a real-time bioreporter. ''Sensors'', 12(1):732-52. | ||
| + | |||
[[http://parts.igem.org/Part:BBa_K801100 BBa_K801100: "RFP-Generator"-Biobrick]] | [[http://parts.igem.org/Part:BBa_K801100 BBa_K801100: "RFP-Generator"-Biobrick]] | ||
| + | |||
[[http://parts.igem.org/Part:BBa_J45014 BBa_J45014:"Banana-Odor"-Biobrick]] | [[http://parts.igem.org/Part:BBa_J45014 BBa_J45014:"Banana-Odor"-Biobrick]] | ||
| + | |||
[[https://2010.igem.org/Team:Groningen iGEM Team Groningen 2010]] | [[https://2010.igem.org/Team:Groningen iGEM Team Groningen 2010]] | ||
| + | |||
[[http://openwetware.org/wiki/IGEM:MIT/2006 iGEM Team Massachusetts Institute of Technology 2006]] | [[http://openwetware.org/wiki/IGEM:MIT/2006 iGEM Team Massachusetts Institute of Technology 2006]] | ||
| + | |||
[[https://2013.igem.org/Team:Queens_Canada/Project/Repel iGEM Team Queens 2013]] | [[https://2013.igem.org/Team:Queens_Canada/Project/Repel iGEM Team Queens 2013]] | ||
Revision as of 15:51, 3 October 2013
Introducing Synthetic Biology Kits at educational institutions
Introduction
Corresponding to this famous statement in pedagogics, the TU Munich iGEM Team of 2013 realized the importance to inspire and teach disciples in educational institutions to long for the endless immensity of Synthetic Biology. We want to make it possible for disciples to take their first steps in the enormously evolving field of Synthetic Biology and let them take a breath of this subject, which will change the world in the near future due to its unlimited fields of application.
The idea is to send an "Educational-Kit" with all required reagents and lab-protocols, based on the experiments we designed, to educational institutions which are interested in inspiring young prospective people to think about a promising career in science. Our experiments are styled constitutive, based on every previously performed experiment, so every disciple, independent of training level, is able to understand and take notice of the resulting phenomena which Synthetic Biology makes possible.
The Educational-Kit
The reagents, which are necessary for the realization of the experiments are all found in the kit, except for competent cells, which have to be stored at -20°C. It was set much value on the autonomous preparation of the final reagents, which have to be used, so nearly all reagents will be send in their blank shape. The aim, the iGEM Team of the TU Munich 2013 is pursuiting, is that the disciples should learn as many steps as possible in the realization of the designed experiments. Following reagents are existent in the kit:
| Reagent | Amount of the Reagent | Is it existent in the kit? |
|---|---|---|
| LB-Medium | Reagents for about 3 L are in the kit | Yes |
| "Banana-Odor"-Plasmid | 100 µl (10 ng/µl) | Yes |
| "RFP-Generator"-Plasmid | 100 µl (10 ng/µl) | Yes |
| "Luciferase"-Plasmid | 100 µl (10 ng/µl) | Yes |
| Isoamylalcohol (>98 %) | 10 ml | Yes |
| Agar plates (Resistance: Kanamycin) | 4 Plates | Yes |
| Agar plates (Resistance: Chloramphenicol) | 2 | Yes |
| L-Arabinose (500 mM) | 3.76 g (50 ml) | Yes |
| Kanamycin | 25 ml | Yes |
| Chloramphenicol | 25 ml | Yes |
| Competent Cells | 6 Tubes (Aliquotvolume: 150 µl) | No |
As you may have noticed, we provide three different plasmids, which are used seperately in each designed experiment. After these experiments have been performed, disciples experienced the whole way from transformation, over cultivating cells, inducing the production of proteins and in the end demonstrate the conversion of a substrate by the produced enzyme.
Experiment Number 1
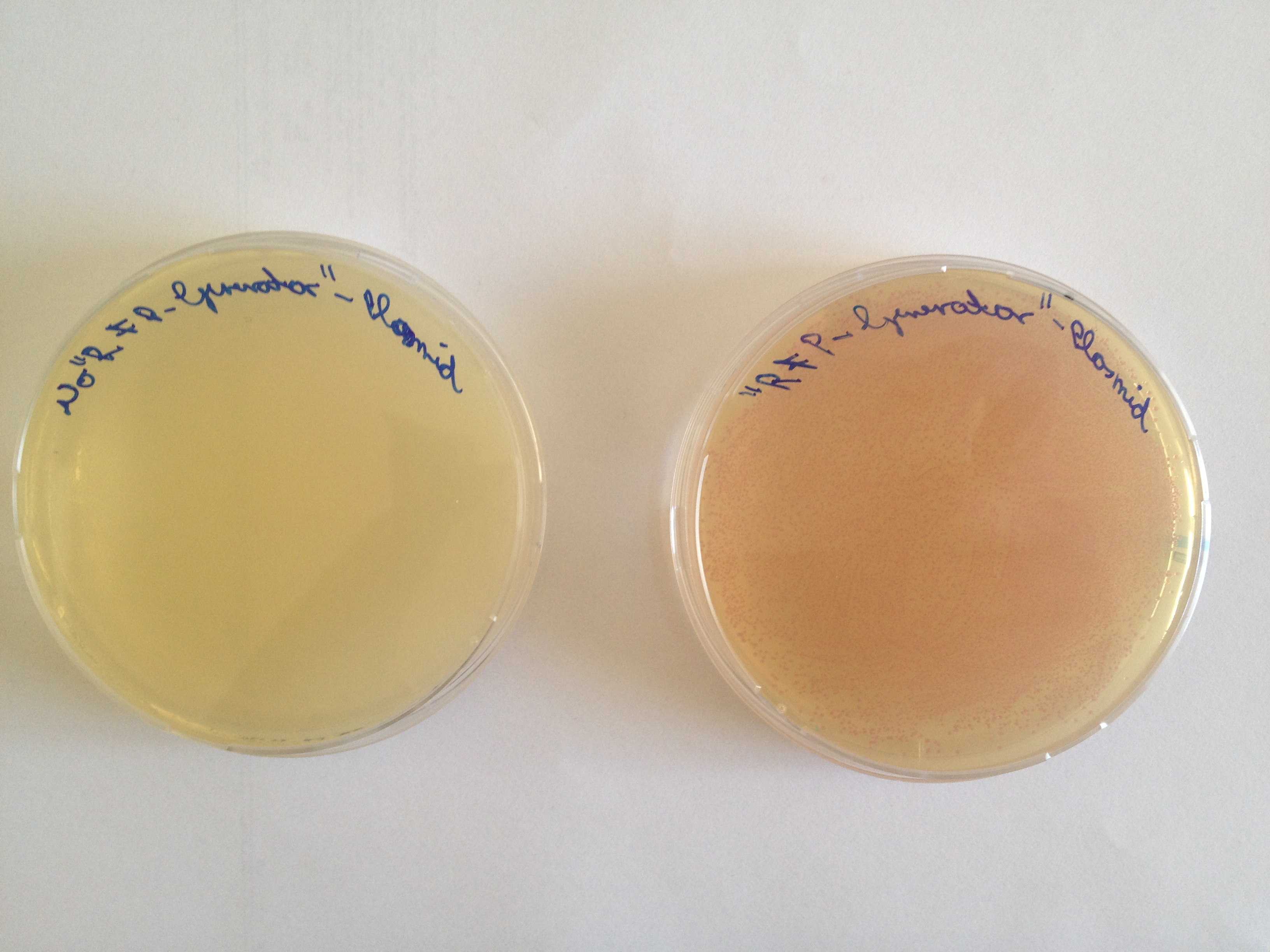
The aim of the first experiment is to transform the [http://parts.igem.org/Part:BBa_K801100 "RFP-Generator"-Plasmid] into Escherichia coli, which contains genes coding for Chloramphenicol resistance plus a red fluorophor, and to plate them on Chloramphenicol-Agarplates. The Agarplates, provided with the antibiotica Chloramphenicol, make a selection process possible, whose aim is to kill all bacteria which have not incorporated the "RFP-Generator"-Plasmid. The expression rate of the red fluorophor is controlled by a constitutive promotor, so the Red fluorescent Protein is expressed continuously. After just about one day of incubation time, the disciples are able to notice a change in colour of the bacteria from ocher to red.
The Biobrick we used to realize this experiment was firstly designed by the iGEM Team of Groningen in 2010 to simplify the cloning step by detecting internalized plasmids, which self-circularized without integration of the desired insert. Bacteria, which internalize self-circularized Plasmids will produce red colonies, so these undesired bacterial colonies can be avoided in the next steps of the experiment, to be sure to use the right construct.
Experiment Number 2
In the second experiment disciples should perform the transformation, learned in experiment one, with the "Luciferase"-Plasmid which encodes genes for Kanamycin resistance and a lux - gene cassettte containing a protein-fluorophor called Luciferase. Another very important point is that the lux-gene is subordinated to a L-Arabinose Operon. After the transformation of the DNA-Plasmid has taken place, discicples should learn how to cultivate bacteria in liquid LB-Medium and how to induce protein expression by using L-Arabinose as an inductor of the L-Arabinose promotor. The glow of the Luciferase can be observed after just a couple of hours.
Experiment Number 3
In the third experiment, all aspects learned in the previous experiments come together. These aspects are necessary to understand the following approaches. The [http://parts.igem.org/Part:BBa_J45014 "Banana-Odor"-Plasmid] encodes an enzyme called Alcohol-Acetyltransferase I. This enzyme is able to convers Isoamylalcohol into Isoamylacetat, which spreads the odor of banana. Additionally there is found a resistance gene for Kanamycin and an inducible L-Arabinose promotor, which controls the expressions rate of the Alcohol-Acetyltransferase I enzyme. The disciples should transform the [http://parts.igem.org/Part:BBa_J45014 "Banana-Odor"-Plasmid] into Escherichia coli, cultivate the successfully transformed bacteria in a bigger scale and in the end, induce the protein expression by using L-Arabinose. In the last step, the disciples should add the substrate Isoamylalcohol to the cell culture, so the produced enzyme can transform it into Isoamylacetat.
The Biobrick, which spreads the odor of banana, after transforming its substrate, was firstly established by the [http://openwetware.org/wiki/IGEM:MIT/2006 iGEM Team of the Massachusetts Institute of Technology in 2006]. Their aim was mainly to produce different compounds in Escherichia coli that smell fragrant. Another very interesting application of the "Banana-Odor" Biobrick is pursued by the iGEM Team of Queens in 2013. This team tries to neutralize foot odour by creating a skin creme containing bacteria with a genetical engineered metabolic pathway to neutralize the volatile compounds that cause these smells. This pathway begins with the uptake of isovaleric acid, a known mosquito semiochemical that is present is foot odour, and converts it into banana smell. By establishing this skin creme, it should be possible to prevent people from being attacked by mosquitos and saved from typical alienable diseases mosquitos spread.
Target Audience
Our target audience have been secondary schools with focus on a biotechnical educational pathway, so all students have access to the required laboratory equipment for the experiments. Before sending the kits to the schools, we tested them in detail - with success. All schools we contacted to spread our Synthetic Biology kits are very enthusiastic concerning our idea of supporting young prospective scientists, so they are very excited to receive and use our kit!
Availability of the School-kit
We appreciate other interested schools in trying our “School kits”. Do not be ashamed of contacting us, so we can supply you with our forward-looking Synthetic Biology “School-kit”!
References
http://www.ncbi.nlm.nih.gov/pubmed/22368493 Close D et al., 2012 Close D, Xu T, Smartt A, Rogers A, Crossley R, Price S, Ripp S, Sayler G (2012). The evolution of the bacterial luciferase gene cassette (lux) as a real-time bioreporter. Sensors, 12(1):732-52.
http://parts.igem.org/Part:BBa_K801100 BBa_K801100: "RFP-Generator"-Biobrick
http://parts.igem.org/Part:BBa_J45014 BBa_J45014:"Banana-Odor"-Biobrick
http://openwetware.org/wiki/IGEM:MIT/2006 iGEM Team Massachusetts Institute of Technology 2006
 "
"







AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351