Team:Paris Saclay/Modeling/FNR
From 2013.igem.org
(→Translate biological knowledges into functional scheme) |
(→Translate biological knowledges into functional scheme) |
||
| Line 33: | Line 33: | ||
=='''Translate biological knowledges into functional scheme'''== | =='''Translate biological knowledges into functional scheme'''== | ||
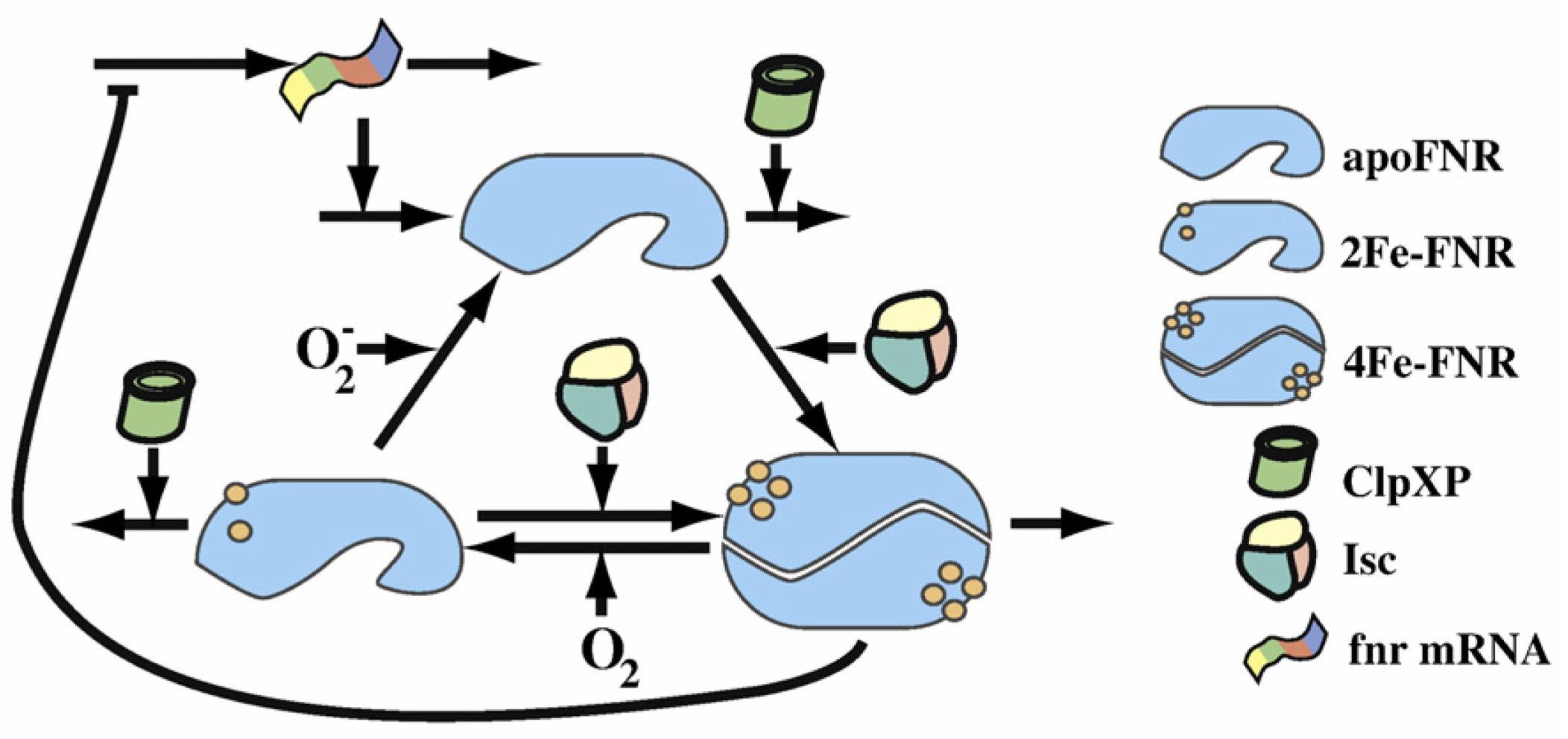
| - | [[File:PSFNRcycle.jpg| | + | [[File:PSFNRcycle.jpg|800px|<center> Tolla and Savageau Regulation of Aerobic to Anaerobic Transitions by the FNR cycle in Escherichia coli</center>]]<br> |
| + | |||
<p>This scheme from the Dean A.Tolla and Michael A.Savageauthe's paper shows clearly the FNR transformation cycle. After the fnr mRNA translation, apoFNR is produced and a dimerization of 2 apoFNR catalyzed by ClpXP forme create the 4Fe-FNR. The 4Fe-FNR can be oxidized by oxygen to 2Fe-FNR then to apoFNR. 2Fe-FNR plus ClpXP can generate 4Fe-FNR. And all 3 FNR derivatives sustain a degradation effect. Those proteins will dissimilate in a while.</p> | <p>This scheme from the Dean A.Tolla and Michael A.Savageauthe's paper shows clearly the FNR transformation cycle. After the fnr mRNA translation, apoFNR is produced and a dimerization of 2 apoFNR catalyzed by ClpXP forme create the 4Fe-FNR. The 4Fe-FNR can be oxidized by oxygen to 2Fe-FNR then to apoFNR. 2Fe-FNR plus ClpXP can generate 4Fe-FNR. And all 3 FNR derivatives sustain a degradation effect. Those proteins will dissimilate in a while.</p> | ||
<br> | <br> | ||
Revision as of 21:47, 4 October 2013
Contents |
FNR oxygen sensor system
As we use Hsim for the modeling, if you want to dig in our model we suggest you to read this page, written by Mr. Patrick Amar, the developer of Hsim.
Roughly speaking, we define a sampling volume, size in nanometer and we declare some molecules with their sizes, their moving speed and their number. Once the simulation starts, those molecules move randomly in the space and trigger some possible reaction between them with some probabilities. There is always somehow deviation from the reality, but we are all excited about this particular simulating modeling.
We represent our FNR regulator model to you by 3 steps. We have chosen HSIM, a multi-agents programme developped at our university in order to follow the assembly, movements and dissociation of a large number of molecules in a virtual cell. HSIM replaces the Ordinary Differential Equations and mimics chemical reactions of the real system by rewriting rules. Following our modeling process, we provided the list of molecules, the rewriting rules, and the initial values of each molecule quantity in a configuration file to HSIM. We also have developed a web version of Hsim which allows user change and test the system parameters.
Translate biological knowledges into functional scheme
This scheme from the Dean A.Tolla and Michael A.Savageauthe's paper shows clearly the FNR transformation cycle. After the fnr mRNA translation, apoFNR is produced and a dimerization of 2 apoFNR catalyzed by ClpXP forme create the 4Fe-FNR. The 4Fe-FNR can be oxidized by oxygen to 2Fe-FNR then to apoFNR. 2Fe-FNR plus ClpXP can generate 4Fe-FNR. And all 3 FNR derivatives sustain a degradation effect. Those proteins will dissimilate in a while.
If we combine the FNR cycle and color reporter system :
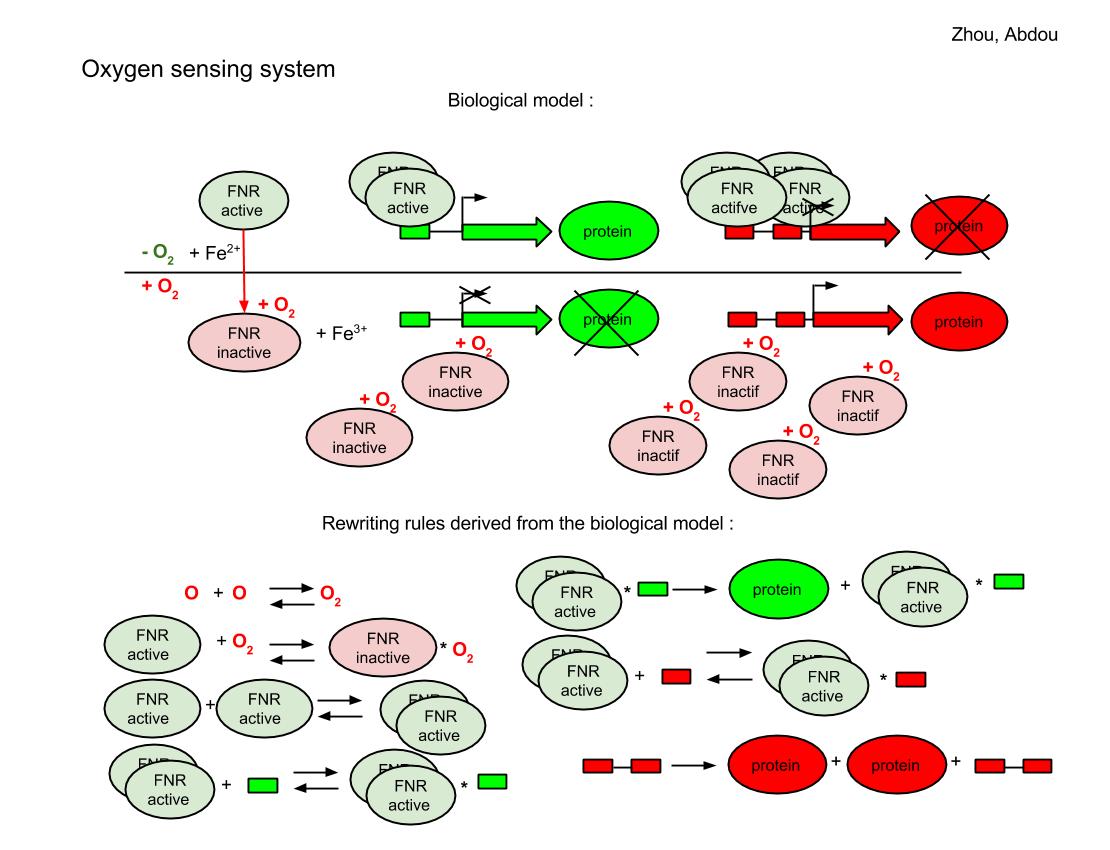
The FNR behave differently for aerobic and anaerobic condition. In the case that oxygen is absent, a pair of 4Fe-FNR fix on the reporter DNA sequence and the green reporter gene is expressed so we will see the green color for those colonies. But a group of 4 4Fe-FNR comes to fix at red reporter’s gene and prevents the expression of red color. If oxygen is sufficient, FNR derivatives keep in inactive form, the red reporter’s gene continue to produce the red color.
Transcribe functional description into equation system
Summary for equations:
- translation of Pfnr gene into FNR protein (FNR without Fe)
- dimerisation of 4Fe-FNR (action of Ise protein)
- oxydation: in aerobic conditions, oxygen inactivates FNR but cell continues to reactivate it (action of Ise protein)
- degradation of each kind of FNR (FNR without Fe, FNR activated, and FRN inactivated), green and red proteins (action of ClpXP
- activation/inactivation of a gene under a repressor promotor (green) and production or not of the cognate protein
- activation/inactivation of a gene under an activator promotor (red) and production or not of the cognate protein
Details
- Translating fnr to FNR
- fnr mRNA generates steadily apoFNR(inactive).
- fnr -> fnr + apoFNR
- Dimerization of apoFNR
- 2 apoFNRs form active 4Fe-FNR(QFeFNRa) with catalyzer Ise.
- apoFNR + apoFNR + Ise -> QFeFNRa + Ise
- Oxidation, from 4Fe-FNR to apoFNR(inactive)
- In aerobical condition, oxygen inactivates FNR, from 4Fe-FNR to 2 inactive Fe-FNR(DFeFNRi)
- QFeFNRa + o2 -> DFeFNRi +o2
- Then, oxidize from 2Fe-FNR to apoFNR
- DFeFNRi + o2 -> apoFNR +o2
- 2Fe-FNR can be reactivate by Ise
- DFeFNRi + DFeFNRi + Ise -> QFeFNRa + Ise
- Degradation FNR
- Protein FNR and its derivation submit a steady degrading rate
- apoFNR + ClpXP -> ClpXP
- DFeFNRi + ClpXP -> ClpXP
- QFeFNRa + ClpXP -> ClpXP
- Activation/inactivation of green gene
- Association between 4Fe-FNR(activated) and green protein gene
- QFeFNRa + GPV -> QFeFNRa_GPV_binded
- Dissociation between 4Fe-FNR(activated) and green protein gene
- QFeFNRa_GPV_binded -> QFeFNRa + GPV
- QFeFNRa_GPV_binded -> PV + QFeFNRa_GPV_binded
- Green protein degradation :catalyzed by ClpXP
- PV + ClpXP -> ClpXP
- Repression of red protein gene : Association between 4Fe-FNR(activated) and red protein gene
- QFeFNRa + GPR -> QFeFNRa_GPR_binded
- Dissociation between 4Fe-FNR(activated) and red protein gene
- QFeFNRa_GPR_binded -> QFeFNRa + GPR
- Association of 4 4Fe-FNR(activated) binded, prevent the production of red reporter protein
- QFeFNRa_GPR_binded + QFeFNRa_GPR_binded -> repression_PR
- Dissociation of 4 4Fe-FNR(activated)
- repression_PR -> QFeFNRa_GPR_binded + QFeFNRa_GPR_binded
- Expression the red gene in aerobic condition
- Production of red protein without repression
- GPR -> GPR + PR
- QFeFNRa_GPR_binded -> QFeFNRa_GPR_binded + PR
- Degradation of red protein
- PR + ClpXP -> ClpXP
Game of parameters
As we mentioned at the introduction, in the interest of time and. In our model Hsim, Kinect of reaction is represented by probability of rules (probability that the reaction takes place). Those probability is quite new stuff for us. In the explication of MR. Amar, those probability is the ratio between the Km of 2 side of chemical equilibrium, however in our model there are lots of no reversal reaction, we got some trouble to deal with them. Units exist dimensions. The article of Dean A.Tolla and Michael A.Savageau show us the relationship among their system, we can peep some reality anyway. For example, we can estimate which reaction is faster than others: the Kinect of oxidation for active FNR is much quick than dimerization of 2 inactive FNR. So what we did is really a game of parameter.
Scenarios and results
Here we prepared 3 scenarios in order to testing our model. Each of them represent one specific condition which could happen in our experiments. The first scenario is about the degradation of red reporter protein in aerobic condition. In the second scenario, the model submit to aerobic then anaerobic condition. The last one is a rare situation, we want to see how our model acts.
In the beginning of each scenario, we let our system run in the distribution mode for few generation, the molecules will spread homogeneously in the virtual bacterium.
- I. Degradation of red reporter protein
In this first scenario, we have a situation that the system has been in a anaerobic condition for a while, there is already some of red protein molecules. What we wish to see is a decline of red protein from initial value to 0, and it is what we observed in our model. There is also a latency due to the time needed for the formation of binding FNR molecules. The synthesis of green protein begins before oxygen running out.
- II. Alternation of reporter protein
In this scenario, we changed the nature of oxygen from “metabolite” to “molecule” which means the quantity of oxygen in our system is no more constant, and distribution of oxygen become heterogeneous. We begin with 5000 molecules of oxygen, and 0 reporter protein. The red reporter protein is synthesized immediately. Oxygen is used to oxidant the FNR active to inactive state, however, when all oxygen is consumed, FNR n-mer begin to synthesize green protein and prevent red one. So we observe a exchange of curve position in the scheme. This scenario reveals the result of our experiments.
Here you can find our configuration files for HSIM: link.
- III. Green reporter protein in aerobic condition
This scenario is a little bizarre, we set at the presence of oxygen, there is still a lot of green reporter in the system. What will happen? It is not quite explicit. So how about a simulation, we amused ourtheves by testing some weird values. We try to find its limit.
And as we can see, after a period of latency, the system has found its way back to the normal situation.
Online simulation
Simple simulation
Complex simulation
Conclusion
Our model reveal successfully the different states of FNR regulation. As you can see, in a anaerobic condition, the number of FNR active is significantly raised, so do the number of green reporter protein. In aerobic condition, because of the presence of oxygen, FNR inactive dominates in the model, the production of green protein ceases so the number of green protein drops, at the same time the repression for the red reporter disappear leading to the restart of the red protein production.
Back to our wet experiments, we used LacZ (blue color BBa_K11550007) and AmilCP (pink color BBa_K11550003) as 2 independent expression reporters which correspond to green reporter protein in our model. And we had not perform the red reporters protein in the wet experiments.
Reference
- Wenmao Meng, Jeffrey Green and John R.Guest FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites Microbiology (1997), 143, 1521-1532
- Parick Amar, Gilles Bernot, Victor Norris HSIM: a simulation programme to study large assemblies of proteins
Article written by Zhou and Damir
 "
"