Team:Leeds/Modeling
From 2013.igem.org
m |
m |
||
| Line 7: | Line 7: | ||
</html> | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| - | {{Team:Leeds_layout|HeaderParky=</div>[[File:Leeds_Parkytopmodel.png|150px|link=|frameless]]<div>|HeaderImage=[[File:Leeds_Modelheader.png|825px|frameless]]|Header=Modelling|content= | + | {{Team:Leeds_layout|HeaderParky=</div>[[File:Leeds_Parkytopmodel.png|150px|link=|frameless]]<div>|HeaderImage=[[File:Leeds_Modelheader.png|link=|825px|frameless]]|Header=Modelling|content= |
We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project. | We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project. | ||
<br> | <br> | ||
Latest revision as of 03:53, 5 October 2013
| We plan to use modelling to help test and characterise our Bio-Devices. This includes modelling our expected fluorescence based upon Fluorescent Protein production, statistical modelling and testing for physical binding versus false positives and various other parts of the project.
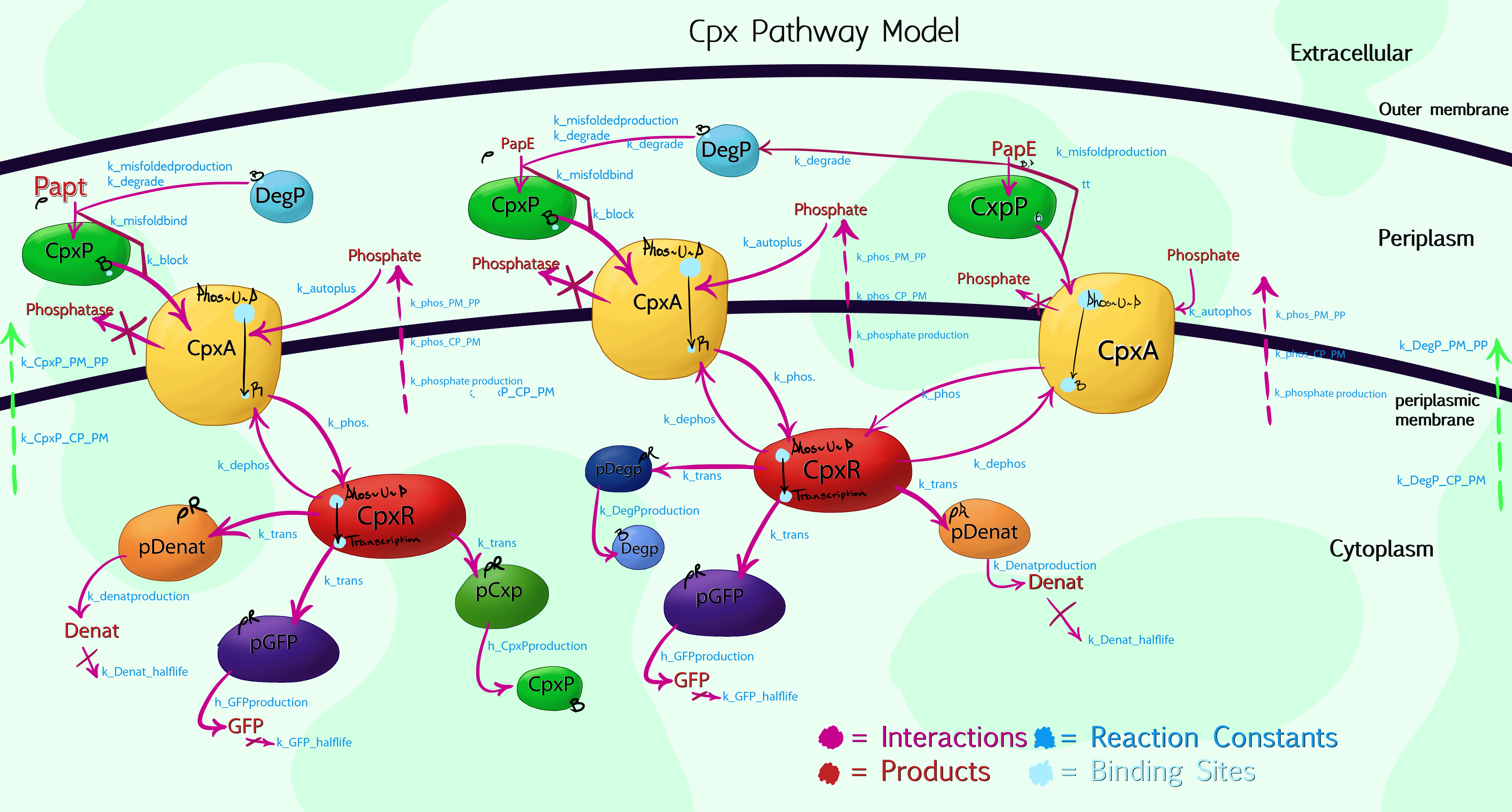
Modelling The Cpx PathwayThe key to making MicroBeagle successful hinges on proper integration with the Cpx pathway. As such, it is essential we develop a god working model, not only to predict how much fluorescence we can expect in different environments, but also to prototype and test potential methods for controlling this; in turn reducing our false positive rate.
Cpx Pathway ModelDue to the high complexity of the Cpx Pathway, it was necessary to graphically plot the processes taking place within the cell by hand, allowing us to then simplify and compartmentalize aspects which, while having an effect on the overall results, were of no interest to us. The above image shows the 3rd iteration of this, with colour-coded interactions, products and proteins. The main enzymes of interest were specifically named, while a general product labelled as 'denaturants' was listed to encompass the more than 100 other Cpx-mediated protein syntheses. This would be inaccurate, if not for the use of Rules-Based software, such as RuleBender - for the model to work, one must list the starting concentrations of each molecular species, hence by setting the 'denaturant' promoter concentration to be at least 100 times greater than that of pGFP or pCpxP, the relative interaction mechanics are preserved. Advice on using the modelFirstly, it is important to bear in mind that the model is an alpha-version, and not fully optimised or debugged. The code is also written expressly in mind of integration with [http://www.ibiblio.org/virtualcell/ VCell] for visualisation purposes, and makes use of the VCell format for compartmentalisation rules - if you do not wish to view the results within VCell, the syntax will need a large re-write. Assumptions / JustificationsThe model gets around the problem of not knowing exactly how every reaction progresses in the patway by making specific assumptions:
| |||||||
 |
| ||||||

| |||||||

| |||||||
 "
"