Team:TU-Eindhoven/Applications
From 2013.igem.org
(→Where and How to Use Our Device) |
JacquesErnes (Talk | contribs) (→Second Application: Tracking of Bacteria in Bacterial Infections Research) |
||
| Line 61: | Line 61: | ||
<big>'''2'''</big> Once in the bloodstream, the CEST contrast producing bacteria travel through it until they find their target. | <big>'''2'''</big> Once in the bloodstream, the CEST contrast producing bacteria travel through it until they find their target. | ||
| - | <big>'''3'''</big> Then a CEST MR | + | <big>'''3'''</big> Then a CEST MR Image is acquired. |
<big>'''4'''</big> The data is analyzed. | <big>'''4'''</big> The data is analyzed. | ||
Revision as of 11:24, 26 October 2013



Contents |
Where and How to Use Our Device
According to different researches, genetically engineered bacteria can be delivered as therapy given that several bacteria, e.g. as Escherichia, Clostridium, and Salmonella, naturally target and accumulate in tumor regions when injected into the bloodstream. LeeBacTumTar Bacteria are able to invade tumors due to the hypoxic micro-environment, poor immune surveillance and increased availability of nutrients.
The most current research being done with bacteria and tumors focuses on inducing cell death within the tumor by means of a cytotoxic agent that is delivered by the bacteria. BugajNextGenTherLeeBacTumTar However there is another potential application for bacteria in what concerns tumors, which is the imaging of the latter. Bacterial-aided delivery of imaging agents has allowed tumor visualization through PET BraderAlEColiTumorDectP. Brader, J. Stritzker, C.C. Riedl, P. Zanzonico, S. Cai, E.M. Burnazi, E.R. Ghani, H. Hricak, A.A. Szalay, Y. Fong et al, Escherichia coli Nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clin Cancer Res 14, 2295-2302 (2008)SogAlPETBactS.A. Soghomonyan, M. Doubrovin, J. Pike, X. Luo, M. Ittensohn, J.D. Runyan, j. Balatoni, R. Finn, J.G. Tjuvajev, R. Blasberg et al, Positron emission tomography (PET) imaging of tumorlocalized Salmonella expressing HSV1-TK. Cancer Gene Ther 12, 101-108 (2004), MRI BenoitAlMRIM.R. Benoit, D. Mayer, Y. Barak, I.Y. Chen, W. Hu, Z. Cheng, S.X. Wang, D.M. Spielman, S.S. Gambhir, A. Matin, Visualizing implanted tumors in mice with magnetic resonance imaging using magnetotactic bacteria. Clin Cancer Res 15, 5170-5177 (2009)DresselaersAlMRIT. Dresselaers, J. Theys, S. Nuyts, B. Wouters, E. de Bruijn, J. Anne, P. Lambin, P. Van Hecke, W. Landuyt, Non-invasive 19F MR spectroscopy of 5-fluorocytosine to 5-fluorouracil conversion by recombinant Salmonella in tumours. Br J Cancer 89, 1796-1801 (2003)HillAlMRIP.J. Hill, J. Stritzker, M. Scadeng, U. Geissinger, D. Haddad, T.C. Basse- Lu¨ sebrink, U. Gbureck, P. Jakob, A.A. Szalay, Magnetic resonance imaging of tumors colonized with bacterial ferritinexpressing Escherichia coli. PLoS One 6, (2011), fluorescence YuAlFluorY.A. Yu, S. Shabahang, T.M. Timiryasova, Q. Zhang, R. Beltz, I. Gentschev, W. Goebel, A.A. Szalay, Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light emitting proteins. Nat Biotechnol 22, 313-320 (2004)ZhaoAlFluorM. Zhao, M. Yang, X.-M. Li, P. Jiang, E. Baranov, S. Li, M. Xu, S. Penman, R.M. Hoffman, Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci USA 102, 755-760 (2005) and bioluminescence ChengAlBiolumC.M. Cheng, Y.L. Lu, K.H. Chuang, W.C. Hung, J. Shiea, Y.C. Su, C.H. Kao, B.M. Chen, S. Roffler, T.L. Cheng, Tumor-targeting prodrug-activating bacteria for cancer therapy. Cancer Gene Ther 15, 393-401 (2008)CroninAlImBiolumM. Cronin, A.R. Akin, S.A. Collins, J.Meganck, J.-B. Kim, C.K. Baban, S.A. Joyce, G.M. van Dam, N. Zhang, D. van Sinderen et al, High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One 7, (2011). The potential of these capabilities lies in the possibility of simultaneously treating and visualizing a specific tumor progression and metastasis in a non-invasive manner. Additionally, targeted fluorescent protein delivery to tumors has been demonstrated to aid surgeons in tumor removal procedures. NguyenAlFluorQ.T. Nguyen, E.S. Olson, T.A. Aguilera, T. Jiang, M. Scadeng, L.G. Ellies, R.Y Tsien, Surgery with molecular fluorescence imaging using activatable cell-penetrating peptides decreases residual cancer and improves survival. Proc Natl Acad Sci USA , (2010) Tumor-specific bacterial delivery of imaging proteins may increase the opportunities of visualizing and treating different types of tumors, with a higher signal-to-noise ratio between tumor and non-tumor tissue. Given that the CEST contrast production from our device is triggered by hypoxic conditions, it could also be possible to link it to bacterial infections considering that hypoxia is also a feature of tissues experiencing a bacterial infection. SawyerOxInfR. G. Sawyer, M. D. Spengler, R. B. Adams, T. L. Pruett, The peritoneal environment during infection: the effect of mono-microbial and poly-microbial bacteria on pO2 and pH. Ann Surg 213, 253-260 (1991)
Inspired by this type of research, we propose two main applications which follow the same idea of targeting a specific region and delivering a compound to it:
1 Tumor imaging based on the sensing of a hypoxic environment.
2 Bacterial tracking in bacterial infection research in animals.
First Application: Tumor Imaging
The idea is to generate the CEST MRI contrast agent on site within the tumor region, facilitating tumor localization in this manner. Considering that the contrast agent will be generated only if the bacteria sense a hypoxic environment, it can be said that using our contrast agent would allow physicians to identify tumors in an accurate way.
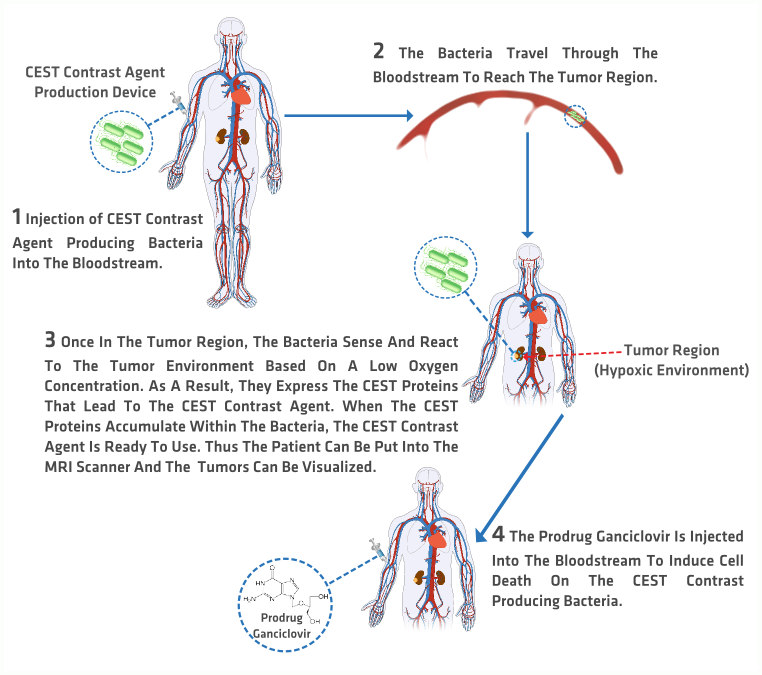
How?
presents a graphic description of the process that would be followed by using the CEST MRI contrast agent to image tumors in humans.
1 A bacterial concentrated solution is injected into the patient.
2 The bacteria in the solution travel through the bloodstream without alerting the immune system.
3 They accumulate in the tumor region due to their ability of sensing hypoxic environments. The anaerobic conditions activate the CEST proteins production. Once the CEST proteins have been produced, the CEST MRI can be taken and analyzed.
4 The bacteria can be disabled and killed by means of activating the kill switch. The switch is enabled by the systemic administration of the prodrug Ganciclovir which induces apoptosis only in the cells that contain the HSV-Thymidine Kinase, in this case the bacteria producing CEST proteins.
Further changes to the current chassis
In order to use our device for this application the current chassis must include two crucial components that enable the bacteria to travel through the bloodstream without raising an alert in the immune system and die when the kill switch is activated.
1 The alteration of its outer surface by exchanging the carbohydrate chains commonly found on it into a carbohydrate layer instead. This alteration will prevent innate and adaptive immune responses, allowing the bacteria to freely travel to the tumor region without being engulfed by phagocytic cells. This solution to the immune system’s response was based on the solutions implemented by the Berkeley iGEM team in 2007 and the research of Dr. J. Christopher Anderson, an assistant professor of bioengineering at the University of California, Berkeley. The Berkeley iGEM team in 2007 proposed to display K1 carbohydrates. K1 carbohydrates are long linear polymers of sialic acid that extend about half the diameter of the bacteria’s surface. Given that polysialic acid is a typical coating on mammalian cells, the human immune system does not recognize K1 as foreign and hence the bacteria are disguised and granted access through the bloodstream. To display K1 carbohydrate in our bacteria it is necessary to also express O antigens, they suggest expressing antigen O16.
2 The chassis must include the HSV-Thymidine Kinase enzyme for the kill mechanism to function adequately.
Second Application: Tracking of Bacteria in Bacterial Infections Research
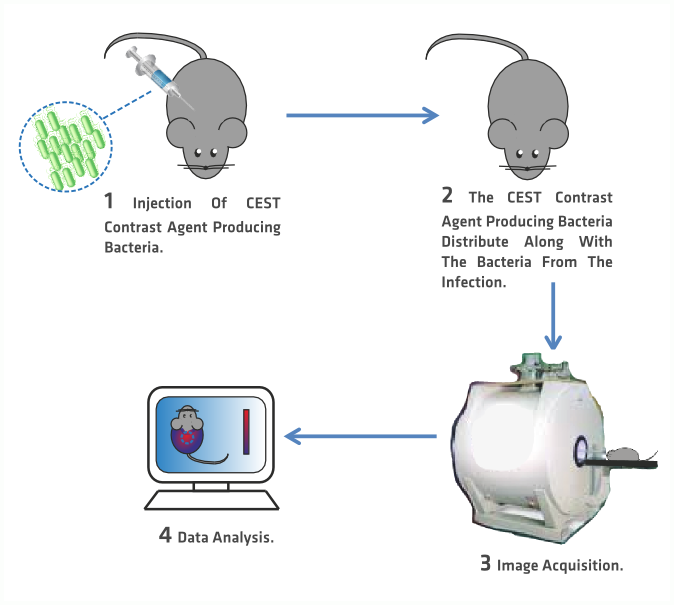
In this application,our device is used as a tool for bacterial infections research, by allowing the researchers to trace the bacteria inside the animal specimen subject to research.
How?
explains the general process that should be followed within this application.
1 The bacterial concentrated solution is injected intravenously.
2 Once in the bloodstream, the CEST contrast producing bacteria travel through it until they find their target.
3 Then a CEST MR Image is acquired.
4 The data is analyzed.
Considering that both applications are similar in principle, no specific model was developed for the second application, but it is possible to modify the parameters of the pharmacokinetics model used in the tumor imaging application to fit the requirements of the second application.
Strengths and Weaknesses of our device in both applications
The ideas we suggest with the two applications previously described take the principles of CEST MRI and delivery systems and apply it to an in vivo production of a CEST contrast agent to enhance the acquired MR images. As any application, what we propose also has advantages and disadvantages. The strongest point of what we propose is the possibility of producing in situ only the required amount of CEST contrast for the image acquisition, summed to the fact that it enables the physician or researcher to easily localize the damaged region. The greatest risk to the success of these applications is the difficulties society might have with coping with the idea of injecting live bacteria into a patient’s bloodstream. This main obstacle can only be surpassed by providing people with the proper unbiased knowledge sources related to synthetic biology and the use of bacteria for clinical and research applications. Therefore, as our human practice project, we have developed an online source of knowledge with links to a variety of papers and other sources related to a specific topic or area of synthetic biology. To know more about it please visit our human practice section in the Wiki.
References
 "
"



