Team:NYMU-Taipei/Modeling/Overview
From 2013.igem.org
| Line 83: | Line 83: | ||
[[Image:NYMU_ molecular way.png|center]] | [[Image:NYMU_ molecular way.png|center]] | ||
| + | |||
| + | (1) sensor(oxyR) model | ||
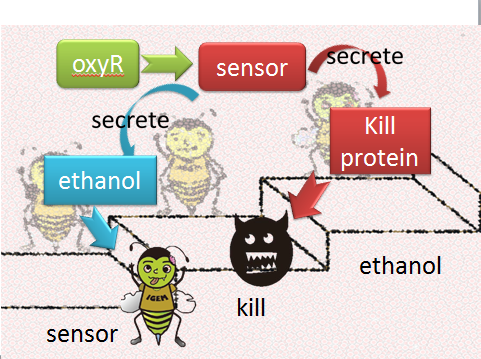
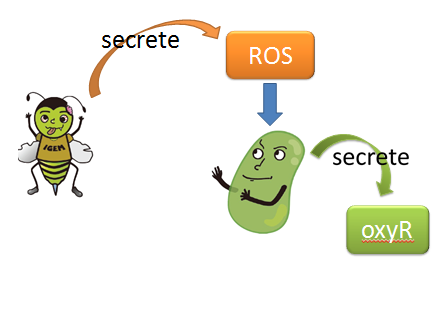
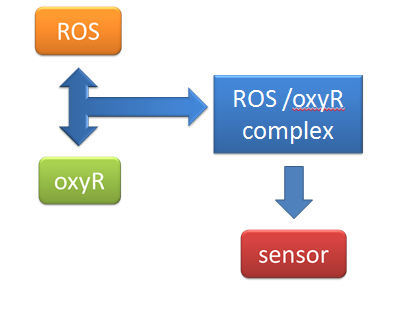
| + | When Nosema enters the bees, it will trigger bees’ ROS (reactive oxygen species) production, which in turn launches Bee.coli’s OxyR production. After that, ROS and OxyR will form a complex and bind to the sensor. As a result, we choose a ROS/oxyR complex-induced promoter as our sensor to activate the whole circuit. (figure1,2) | ||
| + | |||
| + | [[Image:NYMU_ ROSoxyR.png|center]] | ||
| + | figure1: This picture shows that how ROS is produced by the infected bees and trigger the production of oxyR in Bee.coli | ||
| + | |||
| + | [[Image:NYMU_ ROSoxyR complex.png|center]] | ||
| + | figure2: This picture shows that ROS and OxyR will form a complex and bind to the sensor | ||
| + | |||
| + | The aim of this model is to see whether oxyR concentration can reach its effective level to activate the sensor in time. | ||
| + | |||
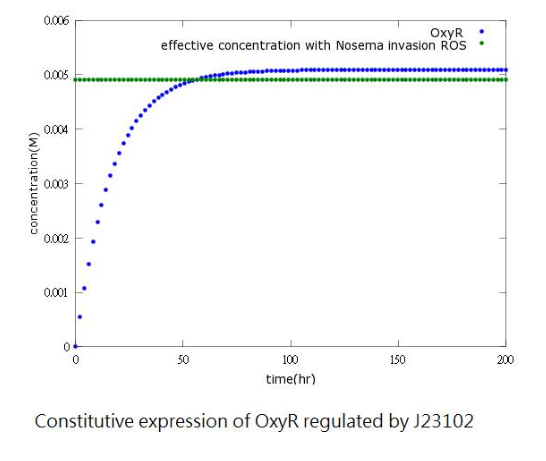
| + | However, since naturally produced oxyR in Bee.coli (our E.coli) is not sufficient, we add a constitutive promoter before the oxyR-producing gene to increase oxyR concentration to reach the effective level. (figure3,4) | ||
| + | |||
| + | [[Image:NYMU_ oxyR concentration to time after adding constitutive promoter.png|center]] | ||
| + | Figure4: oxyR concentration to time after adding constitutive promoter | ||
Revision as of 09:16, 27 October 2013


Contents |
Overview
This year, our team targets to tackle a challenging problem all over the world - CCD, colony collapse disorder by creating a special kind of E. coli. Our project can mainly be separated into four main parts – prevention, sensing and killing, suicuding, and safety.
In the first part prevention, monosidase is used, which can inhibit Nosema polarfilament development. This part is mainly done by experiment.
In the second part sensing and killing, it can further divide into three parts – entrance, sensing, and killing. In entrance part, we use beads (encapsulation) to make it easy for our bacteria getting into the bee. For sensing part, we choose ROS-induced promoters, which can be triggered due to the increase concentration of active transcription factor(OxyR or SoxR). As for killing part, microbial peptides defensin and abaecin are used to pierce Nosema cell wall and then let it be bursted.
Here we are interested in the relationship between concentration ROS, active transcription factor and ROS-induced promoters’ open strength. Furthermore, we also want to know the lag time between sensing the invasion and the production of the killing protein to see if the device can save the bees from being killed by Nosema. As a result, we use sensor model to attain this goal.
However, the spread of E.coli from bee to bee is also another important factor influencing the efficiency of killing Nosema. Consequently, epidemic model is applied to see the relationship between Nosema infection and E, coli treatment.
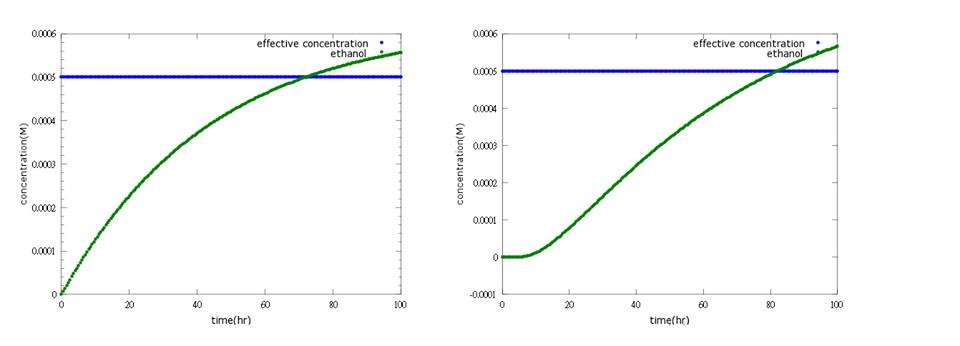
In the third part suiciding, ethanol is used to make bees which are fail to survive after E.coli loses to kill Nosema to suicide itself. Because this part should not be easily opened, otherwise, bees will under the threat of being killed all the time even without the presence of Nosema, we add several terminals behind promoter. Here, ethanol model is used to simulate how many terminals do we need as a threshold.
The last part is safety issue. Since it may be disastrous to the environment if E.coli escapes from bee’s body, we want E.coli to be killed once it leaves bees’ body. Light sensor is used to achieve this goal.
Model highlight 1: PoPS for promoter strength
Mostly, promoter strength is determined by single-round in vitro transcriptions (sequences containing -35motifs, spacer, -10motifs, disc, start, initial transcribed region - bits) like the model of PWMs or in vivo GFP fluorescent assays. However, there are several drawbacks for using such methods to determine promoter strength. For example, promoter strength determined by sequences may be incorrect due to interdependency of motifs [1]; GFP fluorescent assays can only determine the relative promoter strength but the absolute one.
Concerning the disadvantages above, we choose PoPS mechanism for promoter strength for it is closer to the real situation. PoPS is defined as the level of transcription as the number of RNA polymerase molecules that pass a point on DNA each second, on a per DNA copy basis (PoPS = Polymerase Per Second; PoPSdc = PoPS per DNA copy).
Figure1: an example of PWMs [2]
Figure2: an example of GFP fluorescent assays [3]
What’s more, instead of describing transcription and translation together, we separate the two apart by applying the similar mechanism (Ribosome on RBS per second) to translation. The following pictures show the comparison between PoPS combing transcription and translation together and the separating counterpart. (figure3,4). The results conclude that the separate one is closer to real situations and thus is more practical.
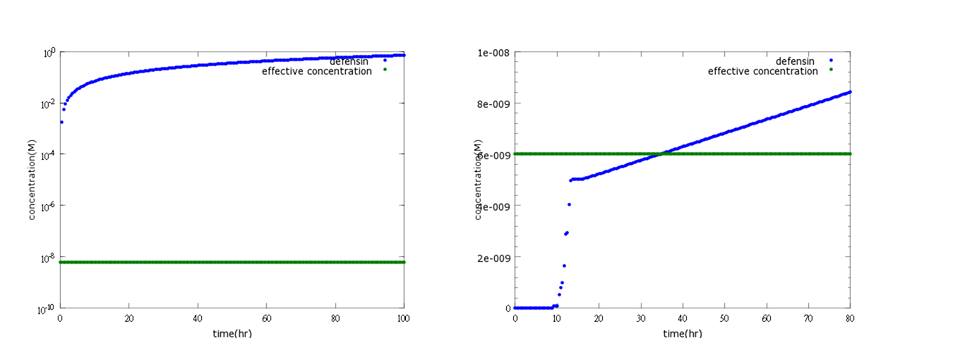
Figure 3: the left shows defensin to time with PoPS combing transcription and translation together; the left shows the separating counterpart. According to experiment, this picture indicates the right one is closer to real situation.
Figure4: the left shows defensin to time with PoPS combing transcription and translation together; the left shows the separating counterpart. The left shows that ethanol production is less affected by terminator addition, while the right one fits the expected result.
Reference: 1. Virgil A. Rhodius and Vivek K. Mutalik. Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, σE. December 29, 2009
2. Virgil A. Rhodius and Vivek K. Mutalik. Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, σE. December 29, 2009
3. John Blazeck, Rishi Garg, Ben Reed, Hal S. Alper. Controlling Promoter Strength and Regulation in Saccharomyces cerevisiae Using Synthetic Hybrid Promoters.
Model highlight 2:How capsule concentration influences Bee.coli survival rate
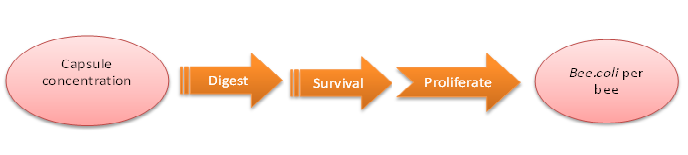
We do this model in order to know whether capsules ingested by bees could be effective enough. That is, whether capsules could be digested by bees’ digestive juice, Bee.coli in capsules is able to proliferate, and finally, Bee. coli could reach the effective concentration (Bee.coli/bee) to defend Nosema infection. (Figure 1.)
The data of capsule digestion rate and Bee.coli survival rate is from experiment, while Bee.coli proliferation rate is assumed according to several papers [1] and confirmed dividing output by input.
The input is capsule concentration and the function contains the transfer of capsule digestion, survival and proliferation of Bee.coli, which then generates the output of Bee.coli/bee.
Last but not least, we retrieve the standard concentration of capsule-contained sugar water which will be fed to be colony via the formula below:
Figure1: model of capsule concentration and Bee.coli survival rate|center
Our project
We divide our project into two main parts, one is molecular way to describe our circuit, including sensor(oxyR) model , kill protein (defensin) model, suicide (ethanol) model; the other is bee-bee interaction for the whole colony.
1. molecular way:
-sensor(oxyR) model
-kill protein (defensin) model
-suicide (ethanol) model
2. bee-bee interaction for the whole colony:
-epidemic model
1. molecular way:
(1) sensor(oxyR) model When Nosema enters the bees, it will trigger bees’ ROS (reactive oxygen species) production, which in turn launches Bee.coli’s OxyR production. After that, ROS and OxyR will form a complex and bind to the sensor. As a result, we choose a ROS/oxyR complex-induced promoter as our sensor to activate the whole circuit. (figure1,2)
figure1: This picture shows that how ROS is produced by the infected bees and trigger the production of oxyR in Bee.coli
figure2: This picture shows that ROS and OxyR will form a complex and bind to the sensor
The aim of this model is to see whether oxyR concentration can reach its effective level to activate the sensor in time.
However, since naturally produced oxyR in Bee.coli (our E.coli) is not sufficient, we add a constitutive promoter before the oxyR-producing gene to increase oxyR concentration to reach the effective level. (figure3,4)
Figure4: oxyR concentration to time after adding constitutive promoter
 "
"