Team:Evry/Sensor
From 2013.igem.org
| Line 9: | Line 9: | ||
<img src="https://static.igem.org/mediawiki/2013/3/38/Iron-sensing.png" align="center"> | <img src="https://static.igem.org/mediawiki/2013/3/38/Iron-sensing.png" align="center"> | ||
</center> | </center> | ||
| - | <br /> | + | |
| + | <br/> | ||
| + | |||
| + | <h2>Construction of the sensor system</h2> | ||
| + | |||
| + | <p> | ||
| + | <i>E. coli</i>'s genome is composed of many Fur binding site. Based on a genome study, we identified 4 promoters which are controled by the FUR protein. | ||
| + | </p> | ||
| + | <ul> | ||
| + | <li>AceB promoter</li> | ||
| + | <li>Fes promoter</li> | ||
| + | <li>FecA promoter</li> | ||
| + | <li>yncE promoter</li> | ||
| + | </ul> | ||
| + | |||
<p> | <p> | ||
We constructed iron-responsive biosensors by combining 3 genetic parts: an E. coli promoter with a Ferric Uptake Regulator (Fur) binding site, a fluorescent reporter (sfGFP), and a transcriptional terminator. These sensors respond to ambient iron by using the <a href="https://2013.igem.org/Team:Evry/Project_FUR">Fur system</a> to repress a target gene. | We constructed iron-responsive biosensors by combining 3 genetic parts: an E. coli promoter with a Ferric Uptake Regulator (Fur) binding site, a fluorescent reporter (sfGFP), and a transcriptional terminator. These sensors respond to ambient iron by using the <a href="https://2013.igem.org/Team:Evry/Project_FUR">Fur system</a> to repress a target gene. | ||
Revision as of 17:31, 28 October 2013
Iron Sensor

Construction of the sensor system
E. coli's genome is composed of many Fur binding site. Based on a genome study, we identified 4 promoters which are controled by the FUR protein.
- AceB promoter
- Fes promoter
- FecA promoter
- yncE promoter
We constructed iron-responsive biosensors by combining 3 genetic parts: an E. coli promoter with a Ferric Uptake Regulator (Fur) binding site, a fluorescent reporter (sfGFP), and a transcriptional terminator. These sensors respond to ambient iron by using the Fur system to repress a target gene.
We constructed 4 differents iron sensor using promoters regions from aceB (BBa_K1163102), fes (BBa_K1163108), fepA (BBa_K1163105) and yncE (BBa_K1163111). Finally, pAceB appears to be the best candidate to build our sensor system. See our results
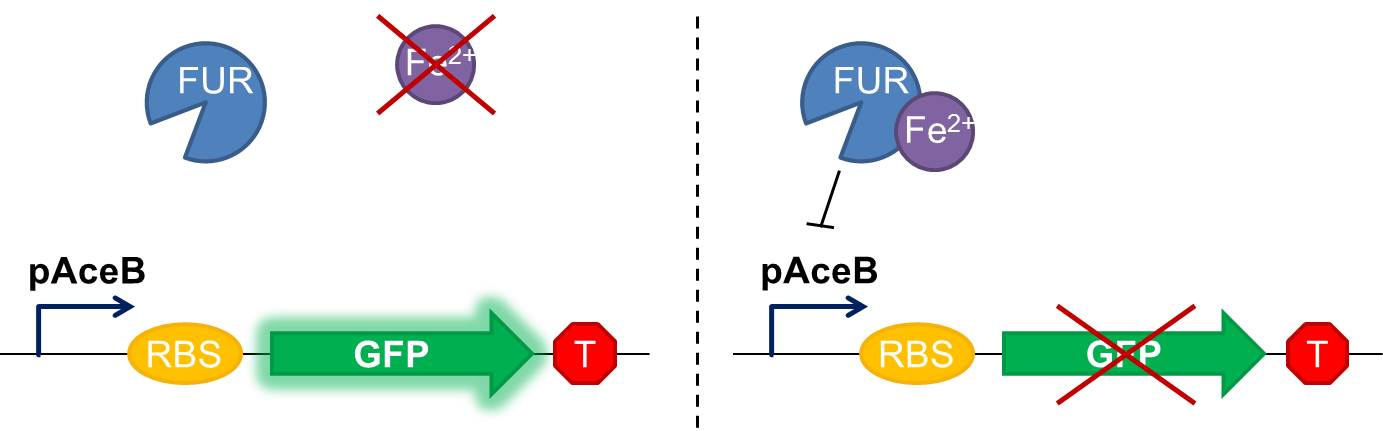
Fig 1 Diagram of our genetic iron sensor. Iron binds the Ferric Uptake Regulator (Fur) to form a complex with high affinity for the Fur box in the promoter, here shown as the aceB promoter. Once the iron-Fur complex is bound to the promoter, it represses transcription of the target gene GFP. GFP expression is thus negatively correlated with iron availability.


Fig 2 Construction of an iron-responsive genetic element by fusing a Fur-regulated promoter with a reporter gene. Promoter-reporter fusions were made with flanking restriction sites that are compatible with Biobrick-based cloning.
| NAME | FIGURE | DESCRIPTION |
|---|---|---|
|
E. coli promoter with Fur binding site |

|
iron-Fur complex binds promoter to repress expression |
|
sfGFP |

|
Fluorescent reporter gene |
|
Terminator |

|
terminator to stop transcription |
|
Plasmid |

|
Biobrick-compatible plasmid backbone |
Table I Genetic elements used to make iron-responsive sensors.
 "
"













