Team:ZJU-China/Project/TheGhostKit/GhostSensor
From 2013.igem.org
The Ghost Kit: Ghost Sensor
Contents |
Background
A universal platform for protein on-site detection
Interest in developing portable detection devices for on-site analysis has risen sharply in recent years. Many areas, including medical diagnostics, environmental toxicology and bioterrorism controls, would benefit from devices that can perform a rapid first-level detection without the need for equipped laboratories or skilled personnel. ZJU-China iGEM team this year successfully built a universal protein detection platform based on the idea of aptamer-induced inner-membrane scaffold dimerization.
In our project, we altogether used two sets of inner-membrane scaffolds as protein sensors. One is based on split GFP and the other is based on split β-lactamase.
Split TEM-1 β-lactamase
Protein fragment complementation assay is a widely used method for protein interaction detection. The two fragments of the protein is rational designed and based on protein interaction assisted folding. TEM-1 β-lactamase is relatively small and monomeric, is well characterized structurally and functionally, can be easily expressed, and is not toxic to prokaryotic cells. Furthermore, no orthologs of β-lactamase exist in many prokaryotes, and thus a protein fragment complementation assay based on β-lactamase could be used universally in prokaryotes without any intrinsic background activity. We split TEM-1 β-lactamase according to [4] into N-terminal consisting of a.a. 26-196 and C-terminal consisting of a.a. 198-290.
Diagram illustrating the split of TEM-1 β-lactamase.
(André Galarneau et al., Nature Biotechnology, 2002, 20, 619-22)
The activity of TEM-1 β-lactamase can be detected with a wide range of methods, such as simple colorimetric in vitro assays using the cephalosporin nitrocefin and assays in intact cells using the fluorescent substrate. There are also many commercial immune colloidal gold strips in China to detect β-lactamase in food and beverage. These strips are cheap, sensitive and robust, making them the ideal candidate for large scale applications.
Design
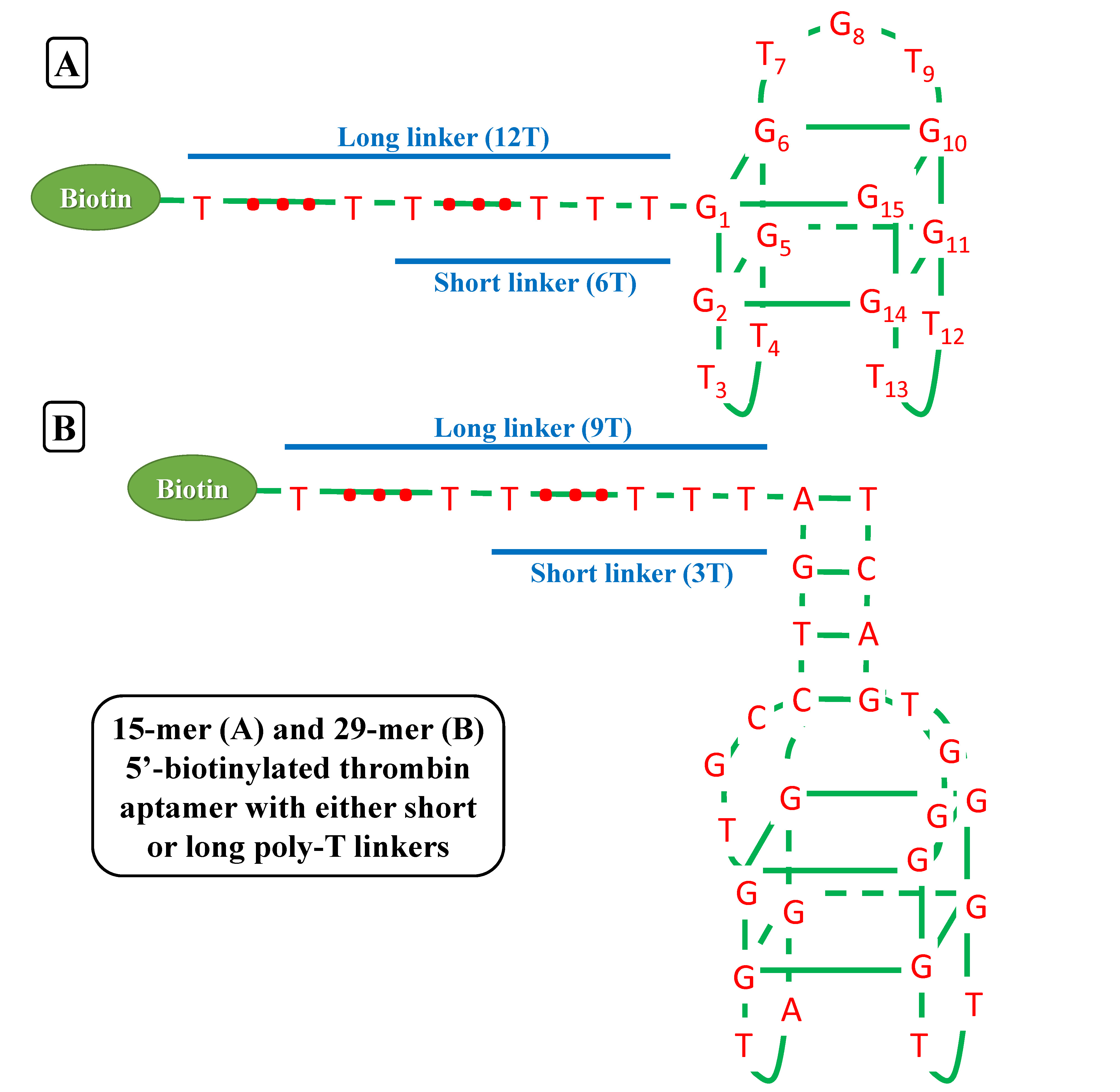
We try to detect thrombin as our proof-of-concept experiment. There have been two well characterized aptamers, one is 15-mer and the other is 29-mer, all show very high and specific affinity to thrombin. We then connected the two aptamers with different length of poly-T linkers, namely short and long linkers, to their 5’ end. At last, all aptamers were modified with 5’ biotin in order that they can bind to streptavidin.
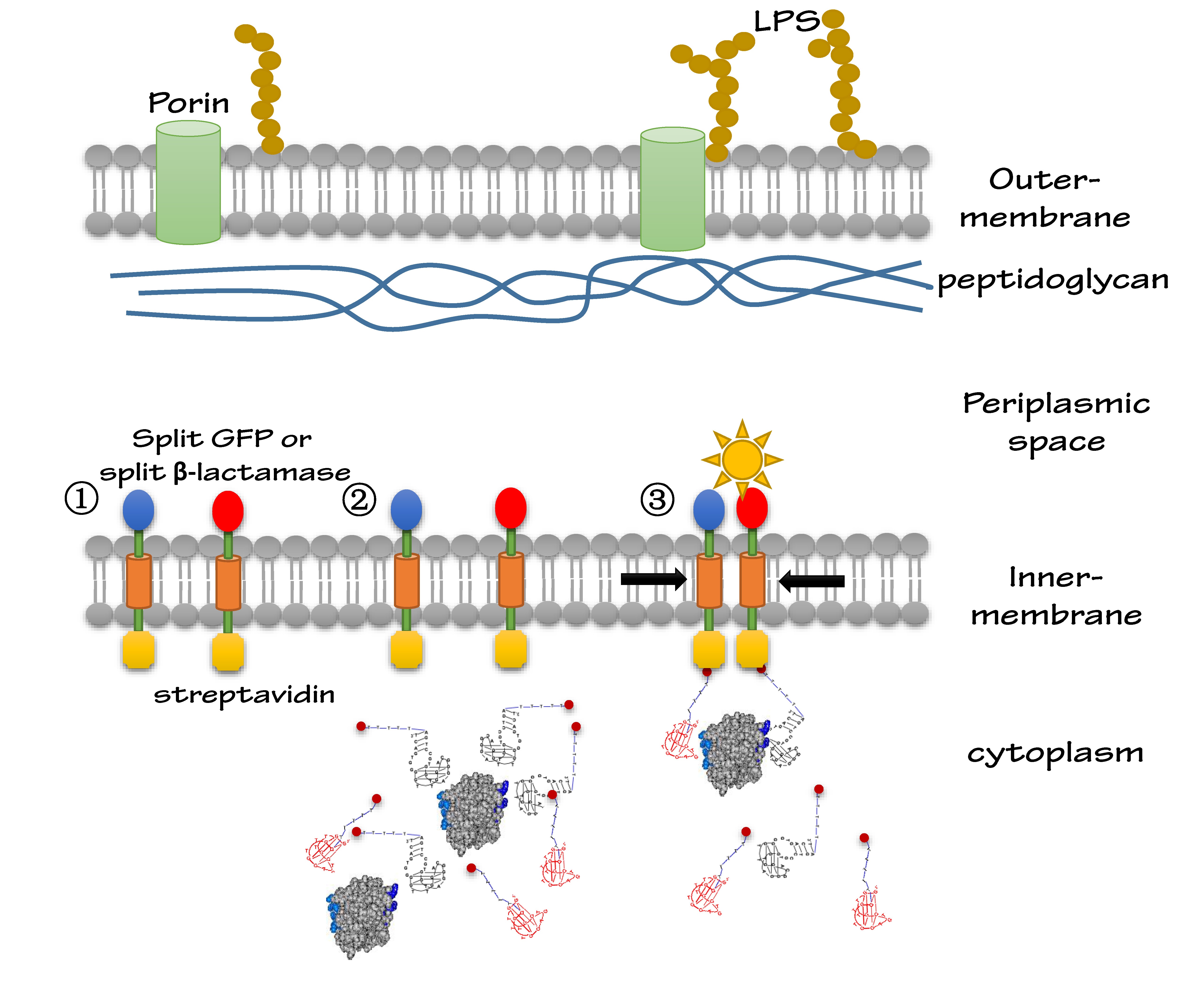
We altogether used two sets of inner-membrane scaffolds, one was based on split GFP and the other split β-lactamase. Our inner-membrane scaffolds consisted of three fusion parts. The split proteins were linked to the periplasmic ends of scaffolds and streptavidin to the cytoplasmic ends. We chose phosphatidylglycerol:: prolipoprotein diacylglyceryl transferase (Lgt) as our transmembrane protein because its function has been well identified by 2012 SJTU-BioX-Shanghai iGEM team. ssDsbA is the signal recognition particle (SRP)-dependent signaling sequence of DsbA. ssDsbA-tagged proteins are exported to the periplasm through the SRP pathway. With ssDsbA fused to the periplasmic terminal, fusion proteins with Lgt are expected to be anchored onto inner membrane of E.coli.
For the first set of membrane scaffolds, two residuals of GFP (BBa_K738006, BBa_K738007) were fused to the periplasmic ends and streptavidin were fused to the cytoplasmic ends. When 19-mer and 25-mer biotin-modified aptamers are added to the solution and thrombin is present, aptamers act like a bridge linking streptavidin-fused membrane scaffolds and thrombin. In this way, two molecules of membrane scaffolds are dragged together and the two residuals of GFP will approach each other in close proximity to become an integrity part capable of giving green fluorescence. With this design, it is believed that we can detect the existence of any protein through fluorescence.
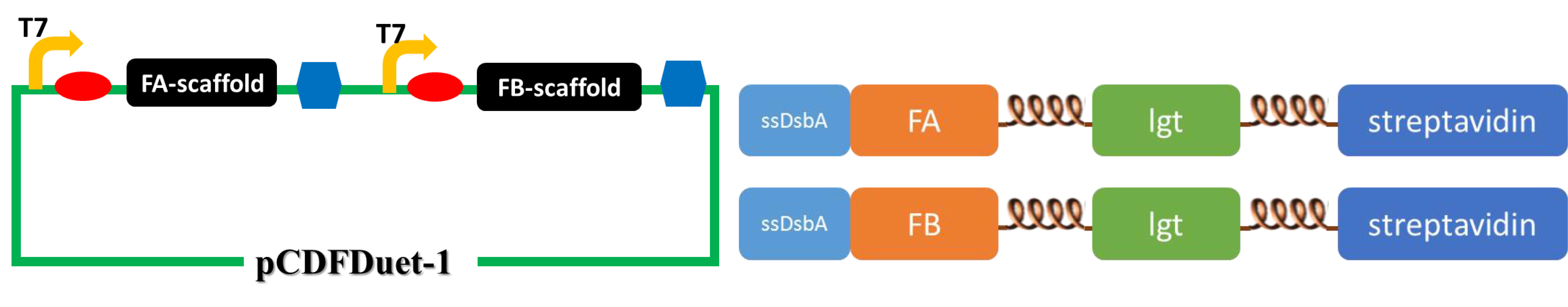
(The upper figure illustrates the construction of the second set of membrane scaffold. We cloned the FA- and FB-scaffold genes into pCDFDuet-1 vector to obtain optimal expression. )
For the second set of membrane scaffolds, we want to see the results with naked eyes instead of fluorescence to expand its application in everyday life. So we fused two residuals of TEM-1 β-lactamase (BBa_K1054010, BBa_K1054011) to the periplasmic ends. Likewise, when 19-mer and 25-mer biotin-modified aptamers are added to the solution and thrombin is present, aptamers act like a bridge linking streptavidin-fused membrane scaffolds and thrombin. In this way, two molecules of membrane scaffolds are pulled together and the two residuals of β-lactamase will meet each other again to become an integrity part with enzymatic activity (in this case, ampicillin resistance).
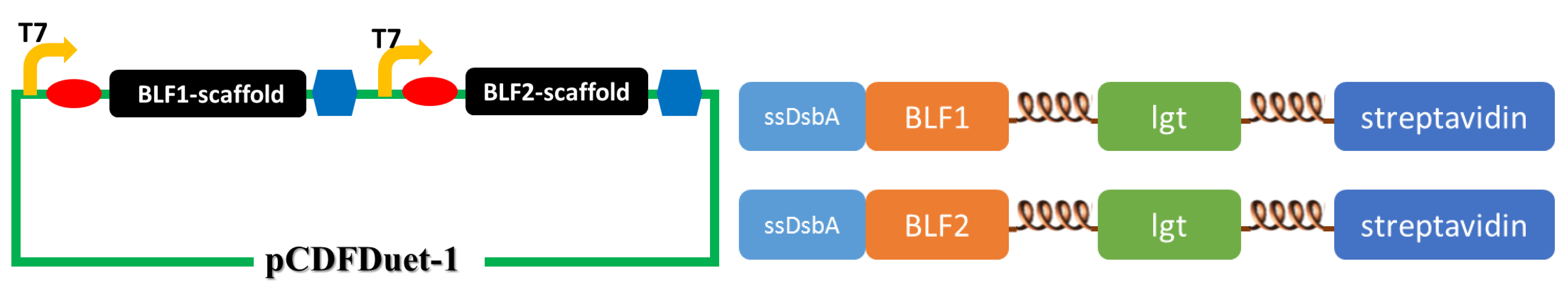
(The upper figure illustrates the construction of the second set of membrane scaffold. We cloned the BLF1- and BLF2-scaffold genes into pCDFDuet-1 vector to obtain optimal expression.)
This activity can be detected by commercial β-lactamase testing strips, which are widely used strips in China initially designed to detect the residence of β-lactamase in drinking milk. One mechanism of these strips are based on immune colloidal gold and we chose it because of its high sensitivity, robustness and relatively low cost.
(The upper figure shows
Results
Detect thrombin using membrane scaffold FA and FB
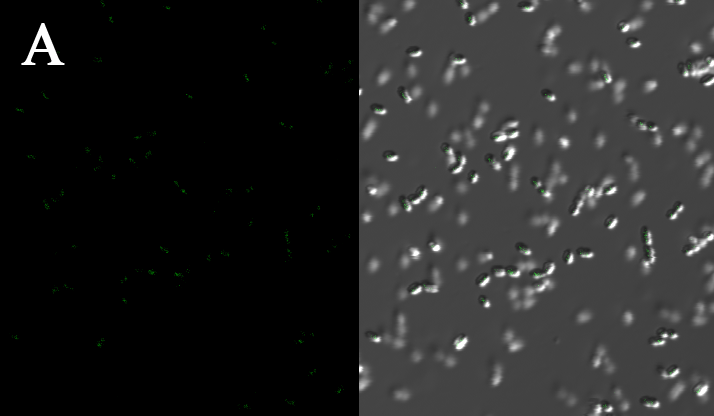
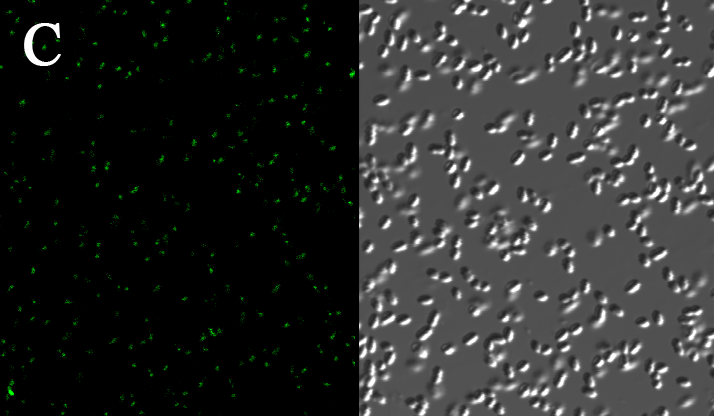
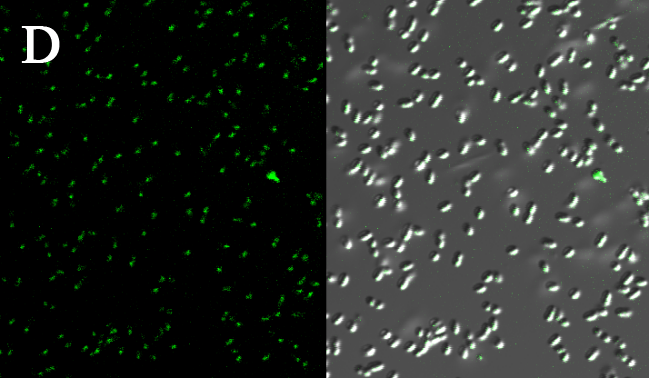
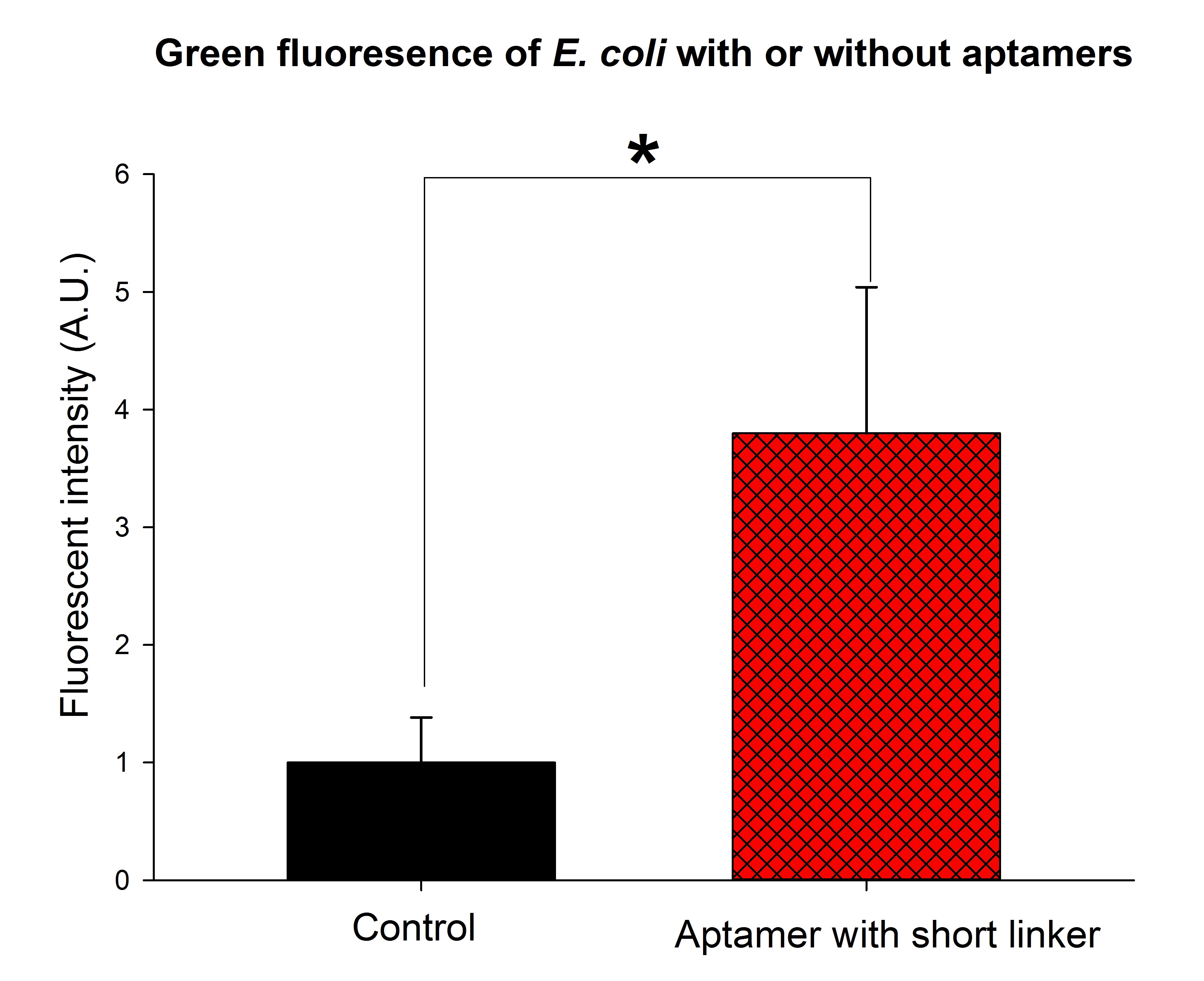
We expressed the first set of membrane scaffolds with split EGFP fragments FA and FB fused to the periplasmic end. Then we tested the function of our membrane scaffolds by adding two sets of aptamers with different linker length. The concentration of thrombin in the solution was adjusted to 0.2nM. 29-mer aptamer and 15-mer aptamer efficiently induced dimerization of membrane scaffolds (Figure C, D versus A) and led to green fluorescence emission under confocal microscopy.
(Fluorescence with or without aptamers. BL21 competent cells were co-transformed with FA, FB and protein E. When the OD-600 value reached 0.4, 0.4% was added to induce the expression of protein E. 2h later cells were harvested and washed two times with PBS. 5µL of bacteria were resuspended in 200 µL PBS, and a final concentration of 0.2nM thrombin was added to the solution. Different aptamers were also included in this point. The reaction system was incubated with shaking in room temperature for 30 min and then green fluorescence was viewed under confocal microscopy. Only very weak green fluorescence was visible when no aptamers were added to the reaction system (A). Biotinylated thrombin can readily induce membrane scaffolds dimerization because there were typically 1-4 biontin in one molecule leading to scaffolds crosslinking (B). 15-mer and 29-mer aptamer with short (C) or long (D) linkers were both effective in inducing crosslinking. But it seemed that long linkers (D) is more potent than short linker (C) in this system. )
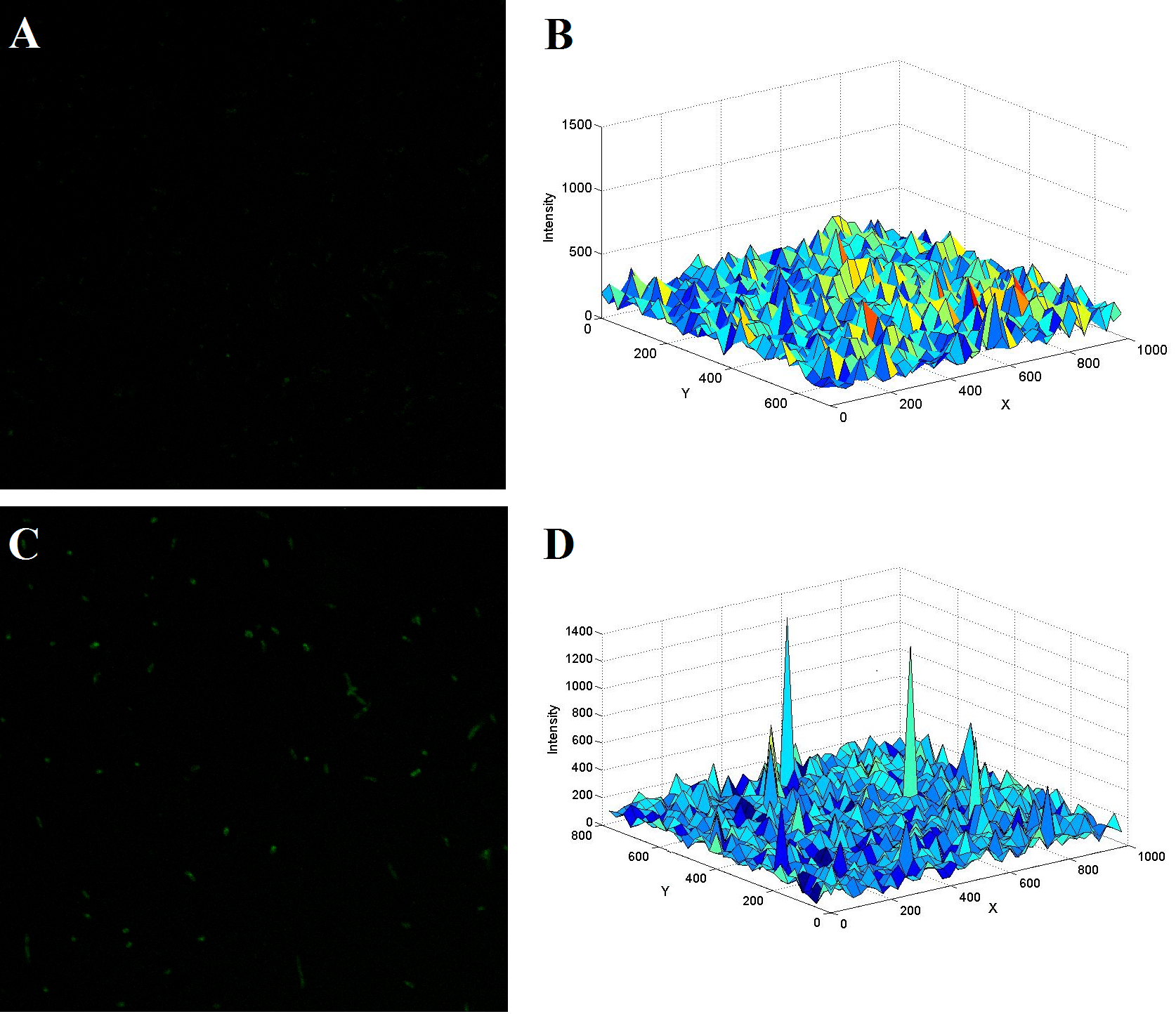
(The upper figure demonstrates another two typical confocal image of E.coli with (C) or without (A) the two kinds of thrombin aptamers. The thrombin concentration was adjusted to a final value of 0.2nM. Figure B and D shows the spatial fluorescent intensity distribution across the image of the left figure accordingly.)
(The green fluorescence intensity was significantly stronger than the control group when thrombin aptamers with short linkers were added to the solution. The final concentration of thrombin was adjusted to 0.2nM. p<0.01, n=3, t-tset. A.U., arbitrary unit.)
 "
"